Influenza
Authors: Julian W. Tang, Nandini Shetty, Tommy T.Y. Lam
Virology
Influenza viruses are members of the Orthomyxoviridae family. They are lipid-enveloped, single-stranded, negative-sense, segmented RNA viruses (Figure 1) that exist in three recognized serotypes: influenza (A, B and C). Whilst influenza B and C are only found in humans, influenza A virus infects various mammals (including pigs and horses, and more rarely, other animals such as dogs) and numerous species of birds (both aquatic and terrestrial). Waterfowl are the natural reservoir of influenza A virus (205). All subtypes of influenza A have been found in waterfowls, and their infections are usually asymptomatic Figure 1A & Figure 1B.
Whereas there is only one type of influenza B virus, influenza A has multiple subtypes, characterized by a combination of the 16 known hemagglutinin (HA) and 9 neuraminidase (NA) genes that code for the viral envelope or surface proteins (205). Of the 16 HA subtypes of influenza A virus, six have been found in a significant number of human infections (H1, H2, H3, H5, H7 and H9); however, so far only three subtypes of HA (H1, H2, H3) and two subtypes of NA (N1, N2) have caused pandemics in humans (Figure 2). In addition, both influenza A and B do exist as several genetically distinct lineages that co-circulate in humans, with the different influenza A strains, in particular, varying sufficiently in the HA component to require different HA antigens to be present in the seasonal influenza vaccine to ensure the induction of adequate specific immunity.
The viral surface proteins (especially HA and NA) play a prominent role in viral entry and escape from the human respiratory epithelial cells within which the virus replicates. Being the major targets of the human immune response, they are known to evolve rapidly under host immunological selection pressure. This generates further novel antigenicity allowing the virus to continually escape from the existing host immunity developed from previous infections or vaccination (88). Such ‘antigenic drifting’ has resulted in new epidemics of influenza A H1N1 and H3N2 in human population about every 3 years, with a similar process in influenza B causing epidemics nearly every 5 years (66).
Reassortment, which recombines genome segments from two influenza strains (when simultaneously infecting the same host cell) into a new progeny virus, is another important molecular mechanism driving the evolution and emergence of novel influenza viruses. Studies have shown that human influenza viruses frequently undergo gene reassortments (92,136), some of which might be advantageous to viral fitness. For example, the abrupt rise of adamantane (i.e. amantadine and rimantadine) drug resistance in H3N2 human influenza during 2004-2006 is attributable to the expansion of an amino acid mutation (S31N) in the viral M2 matrix protein which has propagated via gene segment reassortment involving two distinct lineages of the H3N2 influenza virus (174). De novo combinations of genome segments between human and other animal influenza virus may generate novel virus strains (termed ‘antigenic shift’) with pandemic potential, as these new viruses will not be recognized by previously induced specific host (human) immunity.
Epidemiology
Influenza types A and B cause frequent and occasionally severe diseases in man, with influenza type A being generally more common than type B. The very young (infants) and the elderly are more vulnerable to typical seasonal influenza infections because of their inexperienced or ageing immune system, respectively. In contrast, pandemic influenza can cause mortality in the younger, fitter adult population, which may, paradoxically, result from an over-active immunological response to a completely new pathogen (182) or to a more virulent virus, per se. Sometimes there may be a combination of these effects, such as is seen with 1918 pandemic influenza-like viruses (109) or the more recent avian A(H5N1) influenza virus (144).
Traditional pandemic influenza surveillance has focused on monitoring antigenic shifts, i.e. the more radical changes in the antigenic profile of influenza viruses due to the reassortment of genes between human and zoonotic influenza A viruses. For the more routine epidemic surveillance of seasonal influenza viruses, e.g. the seasonal A(H3N2) and A(H1N1) subtypes, viral isolates are collected throughout the year to keep track of their antigenic drift to determine the most appropriate seasonal influenza vaccine composition for the coming influenza seasons in northern and southern hemispheres (see section on Influenza vaccines below).
Past Influenza Pandemics
During the 20th century, there were three pandemics: the 1918 H1N1 (‘Spanish flu’), the 1957 H2N2 (‘Asian flu’) and the 1968 H3N2 (‘Hong Kong flu’). The ‘Spanish flu’ pandemic coincided with World War I, during which an estimated one third of the world’s population (i.e. around 500 million people) became infected with about a 10% mortality (about 50 million people – though estimates vary quite widely). The subsequent 1957 and 1968 pandemics, which are now thought to have their origins in Asia (7), resulted in a lower morbidity and mortality, yet still had a significant impact on global health. Importantly, these subsequent pandemics supported the hypothesis that pandemics are likely to recur and that there may be a benefit in preparing for them. This belief was reinforced most recently by the emergence of the 2009 A(H1N1) influenza virus – deemed to be the first influenza pandemic of the 21st century. Yet, notably, this virus (also classified as an H1N1 subtype, i.e. as for the 1918 ‘Spanish flu’ virus) has been less pathogenic than those that have occurred previously, with an all-age mortality of about 0.02% (55).
The 1918 Spanish flu was caused by an H1N1 influenza A virus. Although its genome has been sequenced and analyzed thoroughly since 2000 (4,156,157,158,159,183), several hypotheses regarding the origin, particularly whether it had emerged from a pure avian or human influenza virus, or via an intermediate human-avian reassortment virus, still exist and remain controversial (75,76,216). Following the end of the 1918 pandemic, the H1N1 influenza virus continued to circulate in the human population causing recurrent but less severe seasonal outbreaks.
This continued until the emergence of a new subtype, H2N2, in 1957, signaling the beginning of another influenza pandemic. This new H2N2 pandemic strain was found to be a reassortant between avian influenza (HA, NA and PB1) genes and those of the co-circulating human H1N1 virus (which provided the remaining 5 segments: MP, NP, PA, PB2 and NS genes - Figure 2) (106). Relatively soon after, in 1968, it was replaced by another novel reassortant virus classified as an H3N2 subtype, which inherited new genome segments from an avian influenza virus (HA and PB1), which recombined with the contemporary H2N2 human influenza (NA, PA, PB2, MP, NP and NS genes - Figure 2) (106). Since then, this 1968 H3N2 human influenza virus has been circulating globally, and continually mutating, for more than forty years.
Interestingly, another H1N1 influenza virus, which has a highly similar genome to the H1N1 influenza virus that existed just prior to 1957 H2N2 pandemic, resurfaced in 1977. This re-emergent H1N1 virus spread rapidly through the population, leading to an epidemic, the ‘Russian flu’ in 1977-1978 (80). It has remained in the global population and has been co-circulating with the other seasonal influenza virus subtype, H3N2, ever since. Although referred to by some as the ‘Russian flu’ it has not been formally considered as an influenza pandemic virus because it was not completely novel, antigenically, and the disease burden was relatively light. It infected mostly children and young adults because the older population had already been exposed to a similar H1N1 strain that had been prevalent during 1947-1957, giving these older adults significant pre-existing immunity to this more recent H1N1 virus. This ‘Russian flu’ H1N1 virus did not replace the contemporary circulating H3N2 influenza virus, but instead co-circulated with it, with each virus predominating every 2-3 years (66). In fact, the behavior of this ‘Russian flu’ virus, which was not classified as a pandemic virus at the time, has similar aspects to that of the more recent H1N1(2009) virus and there has been some interesting debate about the status of this virus as a true pandemic virus (47).
Historically, it seemed reasonable to expect that new pandemics would emerge from the reassortment between existing human influenza viruses and those of avian origin from Southeast Asia. Therefore, it was something of a surprise when the recent H1N1(2009) pandemic virus appeared to be a reassortant of two swine influenza viruses of distinct genotypes – a Eurasian avian-like swine influenza virus and a triple reassortant swine influenza virus (137,155), emerging from Mexico (Figure 2) (70). What may not have been so surprising for a zoonotic pathogen was that early infections may have been more serious with a lessening of morbidity and mortality as it gradually adapted to man as it spread across the USA then rapidly throughout the rest of the world in just a few months. At present, the pandemic A (H1N1) and the pre-existing seasonal A (H1N1) and A (H3N2) influenza viruses are co-circulating in human population.
Interestingly, ongoing surveillance has reported that each of these subtypes may be predominant in different geographical regions and that this situation is continually changing. Exactly what factors (e.g. climate, herd immunity and vaccine uptake, etc.) drive these differences in predominating, circulating subtypes in different human populations is the subject of some intense research.
Other Zoonotic Influenza Infections of Humans
It is recognized that birds provide a diverse gene pool of influenza A virus from which novel genes may be introduced into human influenza viruses. Although it is now clear that bird-to-human transmission events have significantly contributed to the genesis of previous influenza pandemics affecting the global population, there have been numerous reports of other bird-to-human transmissions affecting individuals. These infections were more sporadic with relatively short transmission chains, usually limited to a single person or at most a small group between whom close contact was routine (e.g. family members).
Six mild cases of H9N2 avian influenza infection in children occurred in Hong Kong in 1999, 2003, 2008 and 2009 (22,23,192). Other cities in China (i.e. Shantou, Shaoguan and Guangzhou) also reported H9N2 human cases in 1998 and 1999 (22). An outbreak of highly pathogenic H7N7 avian influenza in the Netherlands in 2003 caused mostly mild illness in 83 people, but also the unfortunate death of one veterinarian (58).
The most widely reported and widespread human avian influenza cases are those due to infection by the highly pathogenic avian influenza (HPAI) A(H5N1) virus. This is largely due to its extraordinarily high human fatality rates, i.e. around 40-80%, in many Asian countries such as Thailand, Vietnam, Indonesia and China (213). From 2005 onwards, this avian A(H5N1) virus, which originated from China, has spread to the bird populations in some regions of Europe, Middle East and African (16,59,135), mutating and diversifying into various strains or ‘clades’ along the way (208). As of 29 December 2010, there were 512 reported cases of human infection of A(H5N1), with 304 deaths (i.e. a mortality rate of 59%)(213) - and the virus is still circulating in these human populations.
There have also been reports (though far fewer) of zoonotic influenza infections from swine. From 1958-2005, there were 36 laboratory-confirmed cases of human infection by swine influenza viruses (i.e. with swine H1N1 and H3N2 subtypes) in Europe and North America (134). Such human infections by swine influenza (H3N2, H1N2 and H1N1 subtypes) have been reported in Asian countries also, including in Hong Kong, the Philippines and Thailand in 1999, 2004 and 2005, respectively (81,111). Most of these infections were mild, yet there were 6 deaths. Perhaps most famously of all, an outbreak of an A(H1N1) swine influenza virus occurred at Fort Dix, New Jersey, resulting in one death and respiratory illness in up to 230 soldiers in early 1976 (72) – which led to a controversial, and arguably unnecessary, US government-led, mass influenza vaccination campaign (170).
On this basis, future routine surveillance of potential zoonotic origins for the next pandemic influenza virus should be expanded to include swine and perhaps other animals (200).
Clinical Manifestation
Typical Clinical Presentation of Seasonal Influenzas
Clinically, influenza is generally a self-limiting disease starting with an abrupt onset of fever (with or without chills), accompanied by generalized myalgia, rhinorrhea, headache, sore throat and coughing. Gastrointestinal symptoms such as loss of appetite, abdominal pain, vomiting and diarrhea are also commonly reported. The usual reason for admission to hospital is when the infection has spread to the lower respiratory tract, causing a range of pulmonary disease ranging from croup, bronchitis, bronchiolitis to frank pneumonia (203). More rarely, there may be involvement of the central nervous system, including encephalopathy and post-influenza encephalitis, transverse myelitis, Guillain-Barré syndrome and acute necrotizing encephalitis (49,95). Other specific manifestations include myositis, which usually occurs 3 days (range, 0–18 days) after influenza onset (1,94) and myocarditis, though this is a rare complication. In infants, influenza can mimic sepsis more usually caused by various bacterial infections, which can add to the difficulty of a timely diagnosis (45).
The annual epidemics of seasonal influenza now have a well-defined, age-specific mortality pattern, with young infants and the elderly (>65 years old) being most at risk, most probably due to relatively little immunological experience with the virus (in infants) and a waning of specific immunological immunity (in the elderly). Hence, the mortality curve typically appears U-shaped when age-specific excess mortality due to pneumonia and influenza is plotted (93).
Clinical Presentations of Previous Influenza Pandemics
The 1957 ‘Asian’ influenza A(H2N2) and 1968 ‘Hong Kong’ influenza A(H3N2) virus pandemics are now known to have arisen in China. The spectrum of illness caused by these pandemic viruses was similar to other influenza virus infections, though certain individuals, such as those with underlying cardiovascular disease, tended to have severe complications. Similar to seasonal influenza virus infections, the mortality curve during the 1957 and 1968 pandemics was U-shaped, with increased fatality rates, mainly due to various forms of pneumonia, in infants and the elderly (93). Ironically, due to advances in antibacterial therapy during these periods, fatal cases caused by primary influenza viral pneumonia in the absence of any secondary bacterial infections, increased.
Louria et al. (124) reported 33 patients with ‘Asian’ A(H2N2) infection during the pandemic of 1957–1958 of whom 72.7% (24/33) had chronic diseases or were pregnant. Of these, 21.2% (7/33) had leukopenia and the death rate related to acute illness was 27.3% (9/33). Oseasohn et al (142). reported 33 fatal cases due to ‘Asian’ influenza virus infections, describing in particular the rapid death of previously healthy young individuals. They reported Staphylococcus aureus as being the most common co-infecting bacterial pathogen, as well as 39.4% (13/33) having evidence of myocarditis and 12.1% (4/33) diagnosed with encephalopathy.
For the 1918 ‘Spanish’ influenza A(H1N1) pandemic, the most significant difference, as compared to the 1957 and 1968 pandemics, was the unusual W-shaped mortality pattern, with a peak number of excess death amongst the younger adults aged between 20 and 40 years (126). Retrospective analysis of 1918 A (H1N1) cases has revealed a fatal pathological triad of leukopenia, hemorrhagic diathesis and pulmonary edema that killed many healthy young adults during the 1918 pandemic. In addition, extensive non-respiratory system involvement was a notable and unusual feature during the 1918 pandemic (93). These were all features unique to the 1918 A (H1N1) pandemic which were not observed in the subsequent 1957 and 1968 pandemics, nor the more recent 2009 A (H1N1) pandemic.
Surprisingly, there was a less significant excess mortality found amongst the elderly during the 1918 pandemic possibly because this population may have had previous exposures (and therefore a degree of pre-existing, cross-protective immunity) to an even older influenza virus that was antigenically similar to the 1918 A (H1N1) virus. A similar scenario has been postulated with the more recent 2009 A (H1N1) influenza pandemic, during which the younger population were more affected than the elderly (85).
Clinical Features of Human Infections with Avian Influenza A(H5N1) Virus
The 1997 outbreak of avian influenza A(H5N1) virus in Hong Kong was the first confirmed example of a purely avian influenza virus crossing the species barrier into man – apparently without any adaptation in an intermediate host (e.g. the pig). The mortality was over 30% (6 deaths out of 18 confirmed cases) and dramatically alerted the influenza community to the very real possibility of direct zoonotic transfers of animal influenza viruses directly from their animal host to humans (44,220).
Subsequent studies into the nature of various influenza virus receptors in avian and human hosts have led to a greater understanding of the mechanisms by which such species-crossing events can happen (129,172,197), with the recent swine-origin 2009 influenza A(H1N1) pandemic (see below) further emphasizing the significance of this zoonotic transmission route.
The incubation period for avian A(H5N1) influenza virus infection has been estimated to be up to 7 days, but in most cases it is around 2–5 days after the last known exposure to sick or dead poultry (104,190). In cases for which limited human-to-human transmission may have occurred, the incubation period has been estimated to be 3-10 days (191).
Analyses of human influenza A(H5N1) cases in Hong Kong, Vietnam, Thailand and Cambodia have revealed that fever and cough were the most common initial symptoms (46,220). Gastrointestinal symptoms including vomiting, diarrhea and abdominal pain were reported early in the course of illness in some cases (103,190,219). In other patients, pleuritic pain and bleeding from the nose and gums has been observed. Patients with A(H5N1) virus infection were usually hospitalized 4–6 days after onset of illness (191).
Common laboratory findings on admission have included leukopenia, lymphopenia, and mild-to-moderately decreased platelet counts (104,118,190). However, for patients with a clinically mild illness, there was no decrease in the white blood cell count (104). Chest radiographic findings included patchy, interstitial, lobar, and/or diffuse infiltrates, consolidation, pleural effusion, and pneumothorax (6,152). The severe human A(H5N1) infections in the 1997 Hong Kong outbreak were also noted to develop reactive hemophagocytic syndrome which was a unique pathologic feature in 3 of the 6 fatal cases, as were increased blood levels of interferon-α, tumor necrosis factor- α and other cytokines. This suggests that cytokine responses contribute significantly to the pathogenesis of human A(H5N1) infections (146,188).
So far, in the fatal cases of human influenza A(H5N1) infection, the median time from onset to death has been about 9 days (191). The fatality rate amongst hospitalized patients was high and varied widely between countries (33–100%), although the overall rate is probably much lower because it has been difficult to estimate the number of mild or asymptomatic human A(H5N1) infections in the community (8). Acute respiratory distress syndrome (ARDS) complicated 76.5% (13/17) of the more recent avian A(H5N1) cases in Thailand, and 44.4% (8/18) of cases in the earlier 1997 Hong Kong outbreak. Multiple organ failure with signs of renal dysfunction with or without cardiac compromise was often observed. Whether the severity of illness varies by clade or subclade of A(H5N1) virus infection, by age, or by immunological, genetic, or other factors is as yet unconfirmed (191).
Most fatal cases of human A(H5N1) did not have any history of pre-existing disease, which contrasts with the usual pattern of predisposing factors leading to severe and fatal cases of seasonal influenza infection that occurs in annual epidemics during inter-pandemic periods. Yet, in A(H5N1) -infected patients who did have underlying cardiovascular, pulmonary and renal diseases, severe illness was also more common (8).
Pandemic Influenza 2009 A(H1N1) Virus Infection
Brief History
In late March 2009, an outbreak of a respiratory illness emerged in Mexico. This was soon shown to be caused by a novel swine-origin influenza A(H1N1) virus (S-OIV), initiating the first influenza pandemic of the 21st century (147).
Most illnesses caused by the 2009 A(H1N1) virus have been acute and self-limited, with the highest attack rates reported among children, young adults and pregnant women (160,161). Adults older than 60 years of age were spared relative to the younger age groups presumably because of prior exposure to antigenically related influenza viruses earlier in life, resulting in the development of cross-protective antibodies (55,85,131).
In the UK, reported case fatality rates were 26 (range 11-66) per 100 000. Interestingly, it was lowest for children aged 5-14 (11, range 3-36, per 100 000) and highest for those aged >or=65 (980, range 300-3200, per 100 000). In the 138 people in whom the confirmed cause of death was pandemic A(H1N1), the median age was 39 (interquartile range 17-57) (55). These figures are not too dissimilar to those in the United States, where the case fatality rate for symptomatic illness was estimated to be 0.048% (i.e. 48 per 100,000), being highest in persons aged 18 years and older, and lowest in children aged 5-17 years (150).
In contrast to seasonal influenza most of the serious illnesses caused by the pandemic virus occurred amongst children. In the UK 13% of hospitalized patients were admitted to a high dependency or intensive care unit and 5% died - 36% of these deaths were in patients under the age of 16 years. Non-white and pregnant patients were over-represented and 45% of patients had at least one underlying condition, mainly asthma (138). Young children appear to be particularly vulnerable to the 2009 A(H1N1) virus infection. They present with marked irritability, severe lethargy, poor oral intake, dehydration resulting in shock, and seizures (84,121). Complications of influenza included invasive bacterial coinfections, encephalopathy or encephalitis (sometimes necrotizing), and diabetic ketoacidosis (115,140). Respiratory transmission from a symptomatic mother to a newborn was reported and newborn infants manifest with apnea, tachypnea, cyanosis, and lethargy (5).
Other pre-existing comorbidities on admission with the 2009 A(H1N1) infection that were associated with poor clinical outcomes included obesity and pulmonary conditions other than asthma or chronic obstructive pulmonary disease (COPD). In addition, radiologically-confirmed pneumonia and a raised C-reactive protein (CRP) level (≥100 mg/L) were associated with severe illness. Overall, of all inpatient deaths in this UK study, 59% occurred in previously healthy people (138,161). The Spanish experience also confirmed that pregnancy as well as obesity, and previously healthy individuals, were at risk of dying.
In the United States, among patients who were hospitalized with pandemic influenza, 32-45% were under the age of 18 years, with approximately 9-31% requiring admission to an intensive care unit (ICU) (99). Approximately 25-50% of patients with 2009 A(H1N1) virus infection who were hospitalized or died had no reported coexisting medical conditions. Underlying conditions that were associated with complications from seasonal influenza were also risk factors for complications from 2009 A(H1N1) virus infection (160). A report by a consortium of Australian and New Zealand intensive care physicians stated that as many as one-third (n=68) of patients on their ICUs received extra-corporeal membrane oxygenation (ECMO). These ECMO-treated patients were often young adults with severe hypoxemia and despite the use of ECMO still had a 21% mortality rate by the end of the study period (52).
It has been found that pandemic influenza A(H1N1) virus infections can be life-threatening for pregnant women, particularly for those in the third trimester (19). In pregnant women delivering before 37 weeks gestation, almost all had pneumonia and approximately 50% of the women in one case series did not have an additional risk factor other than being pregnant (89). Although pregnant women represent only 1 to 2% of the population, among patients with 2009 A(H1N1) virus infection, they accounted for up to 7-10% of hospitalized patients, 6-9% of ICU patients and 6-10% of patients who died. There appears to be a particularly increased risk of death among infected women during the third trimester, especially among those who have coinfection with the human immunodeficiency virus (HIV) (5).
Clinical Presentation
The incubation period is approximately 1.5-3 days, similar to that of seasonal influenza. Patients usually become symptomatic within a week of exposure and are infectious for about 8 days thereafter (25,53). Infection with the pandemic influenza A(H1N1) virus causes a broad spectrum of clinical syndromes, ranging from afebrile upper respiratory illness to fulminant viral pneumonia. Most patients typically present with fever (95%), cough (88%), shortness of breath or dyspnea (60%), fatigue (43%), runny nose (38%), sore throat (31%), headache (34%), and myalgia (36%) (99). A proportion of affected patients may present with gastrointestinal symptoms such as diarrhea and vomiting. A small subset of patients have presented solely with gastrointestinal or neurological symptoms without fever or cough (53,99). Systemic symptoms are frequent. Gastrointestinal symptoms (including nausea, vomiting, and diarrhea) occur more commonly than in seasonal influenza, especially in adults (53).
In one large US study, the most common presenting symptoms of patients admitted to intensive care with pandemic A(H1N1) included fever in 97%, cough in 93%, shortness of breath in 87%, fatigue in 46%, vomiting in 25%, and diarrhea in 24% (99). Dyspnea, tachypnea in children, chest pain, hemoptysis or purulent sputum, prolonged or recurrent fever, altered mental status, dehydration, and reappearance of lower respiratory tract symptoms after improvement were all signs of progression to more severe disease or complications (112,204). The principal clinical syndrome leading to hospitalization and the need for intensive care was a diffuse viral pneumonitis associated with severe hypoxemia, ARDS, or in some cases, sometimes shock and renal failure. This clinical syndrome accounted for approximately 49-72% of ICU admissions during the A(H1N1) pandemic (112,204). It is important to note that currently available prognostic algorithms for community-acquired pneumonia, such as CURB-65 (a measure of confusion, urea nitrogen, respiratory rate, and blood pressure and an age of 65 years or older), may not apply (5). In addition, there have been sporadic cases of pandemic influenza A(H1N1) infections with neurologic manifestations, presenting as confusion, seizures, unconsciousness, acute or post-infectious encephalopathy, quadriparesis and encephalitis (140). Particularly in children, this virus seems to have a propensity to cause neurologic complications (3,34,60). Influenza is not one of the viruses traditionally considered as a cause of neurologic illness in children, but this particular pandemic A(H1N1) virus subtype may well become part of the routine differential diagnoses in such pediatric presentations, if this virus starts to co-circulate in human populations, long-term, as one of the seasonal influenza viruses.
Radiographic findings commonly include diffuse mixed interstitial and alveolar infiltrates (see case studies), although lobar and multilobar distributions occur, particularly in patients with bacterial coinfection. Chest computed tomography has shown multiple areas of ground-glass opacities, air bronchograms, and alveolar consolidation, particularly in the lower lobes (99).
Secondary Bacterial Infections with Influenza Viruses
It is now well-accepted that seasonal and pandemic influenza are frequently complicated by bacterial infections (217). Secondary bacterial respiratory infections can be either combined viral/ bacterial pneumonia or post-influenza pneumonia, which differ in their pathogenesis. During combined viral/ bacterial infection, the virus, the bacterium and the host interact with each other. Post-influenza pneumonia may, at least in part, be due to resolution of inflammation caused by the primary viral infection. These mechanisms restore tissue homeostasis but greatly impair the host response against unrelated bacterial pathogens (195).
Secondary bacterial pneumonia is a common cause of death in persons with seasonal influenza and co-infections have been found with about 25% of all influenza-related deaths. Of laboratory-confirmed cases of community-acquired pneumonia, about 30% involve bacterial–viral co-infection. Streptococcus pneumoniae is the most common cause of community-acquired pneumonia and bacterial co-infection with influenza A (83).
Staphylococcus aureus is also a significant bacterial co-pathogen. In a retrospective, descriptive study of all reported influenza-associated pediatric deaths in the 2007–2008 influenza season in the United States, it was found that S. aureus was the most common bacteria isolated from children with influenza-associated mortality. In 85% of these children, specimens yielding S. aureuswere obtained within three days of inpatient admission. S. aureus isolates were associated with older age and lack of high-risk medical conditions (143).
During the 2009 A(H1N1) influenza pandemic, secondary bacterial pneumonia, usually caused by S. aureus (often methicillin-resistant MRSA), S. pneumoniae and Streptococcus pyogenes was found to occur in 20 to 24% of ICU patients, and was identified in 26 to 38% of patients who died, often in association with a more rapidly deteriorating clinical course, with death occurring within 2 to 3 days in some cases (5).
The US Centers for Disease Control and Prevention (CDC) examined post-mortem lung specimens from patients with fatal cases of pandemic influenza A(H1N1) for bacterial causes of pneumonia. Of the 22 cases with histopathologic, immunohistochemical and molecular evidence of co-infection with identified bacteria, there were 10 cases with S. pneumoniae, 6 with S. pyogenes, 7 with S. aureus, 2 with S. mitis, and 1 with Haemophilus. influenzae; 4 cases involved multiple pathogens (28).
Laboratory Diagnosis
The traditional method of identifying influenza virus in clinical samples has been culturing the virus in embryonated chicken eggs (Figure 3A) or more recently, in permissive cell-lines such as primary rhesus monkey kidney (RhMK) cells or continuous epithelial cell lines such as Madin-Darby Canine Kidney (MDCK) cells (Figure 3B). After 1-10 days, the presence of any influenza virus can be confirmed with specific immunofluorescence testing with hemagglutination being a useful (but non-specific – since other non-influenza viruses such as rubella may also cause hemagglutination) marker of virus growth.
Although viral culture is highly sensitive and specific and is one of the few remaining ‘catch-all’ techniques (the other being electron-microscopy) with the advantage of amplifying the virus for further characterization, it is relatively slow. Since it may take several days for the culture to become positive, the results may not be ready early enough to inform the immediate clinical management of the patient. Hence, though viral culture may still be retained in virology reference laboratories as the ‘gold standard’ for viral diagnosis, in hospital diagnostic laboratories, the use of the reverse-transcription polymerase chain reaction (RT-PCR) has become more popular due to its increased speed (i.e. a decreased turnaround time).
In recent years, a variety of molecular approaches to direct detection of viral nucleic acids in clinical specimens have also been explored for rapid diagnosis, including nucleic acid hybridization as well as RT-PCR, though PCR is increasingly becoming the most widespread technique (Figure 4). In particular, it has the advantage of potentially being more sensitive [sensitivity = no. of true positives/ (no. of true positives + false negatives)] than cell culture, and is able to detect the presence of virus nucleic acid in samples in which the virions have lost viability.
The converse of this is that RT-PCR testing, by itself cannot determine the viability of the virus even though its RNA is detectable. Although viability is not unreasonably assumed if the viral RNA is detected in a clinical specimen recently obtained from a live patient, for environmental and infection control studies where the viability of the virus detected on surfaces is important, RT-PCR testing cannot help and viral culture is still required – to demonstrate viability and the potential for onward transmission. For more highly pathogenic influenza virus subtypes, e.g. avian influenza A(H5N1), PCR techniques are safer as the live virus should only be handled under appropriate biocontainment (enhanced BSL-3 or higher) conditions.
Another advantage of the diagnostic PCR assay is that it can be ‘multiplexed’ to allow the detection of many different viruses within the same family (e.g. influenza A types H1, H3, H5, H7 and H9 viruses), or which cause a similar clinical syndrome, e.g. a ‘respiratory virus panel’ which includes influenza, parainfluenza, respiratory syncytial virus, rhinoviruses, coronaviruses and adenoviruses, all of which cause respiratory infections in humans. There are many commercial kits available for such testing, but they can only be applied effectively in professional diagnostic laboratories with trained, skilled manpower and the required equipment.
With the recent pandemic influenza A(H1N1) pandemic virus, specifically designed PCR assays became the preferred method of detection as the pandemic virus was found to grow more slowly than seasonal influenza virus in conventional cell culture and it is less easily detected by the conventional immunofluorescence tests that are usually quite sensitive for seasonal influenza A. In fact, throughout the pandemic (June 2009- August 2010), PCR methods were the preferred method of detection of this novel virus mainly because these conventional methods were insufficiently sensitive to diagnose new human cases (33,77).
Rapid (Point-of-Care) Test Kits for Influenza
As a result of the massive numbers of individuals presenting to community and hospital healthcare services, annually, seasonally, with influenza-like illness (ILI) each year, there has been considerable interest in the application of rapid, point-of-care (POC) tests for influenza diagnosis for several years. Such test can offer almost instant results in busy community healthcare clinics, when sending samples to the nearest diagnostic laboratory can be time-consuming and costly, and may reduce the unnecessary prescription of broad-spectrum antibiotics for common respiratory virus infections.
These tests were originally developed for the rapid diagnosis of the seasonal influenzas A and B and include such kits as QuickVue (Quidel Corp., San Diego, CA – see Figure 5), Binax NOW (Binax Inc., Scarborough, MA) and Directigen Flu A+B and/or Directigen EZ Flu A+B (Becton Dickinson, Sparks, ME), all of which are licensed for this purpose in the USA. Some of the kits on the market cannot distinguish between influenza types A and B, and none of them can distinguish between the influenza A subtypes H1, H3 and H5.
For most of these kits, their reported sensitivities for detecting seasonal influenza viruses range over 30%-95%, depending on the conventional laboratory ‘gold standard’ test against which they are compared as well as the care and experience of the operators. Nearly all of them work on the same principle of immunochromatography, i.e. the detection of viral antigen in respiratory secretions. In these tests, a sample of respiratory secretions is treated with a mucolytic agent and then tested, either on a filter paper, in an optical device, or with a dip-stick in which reaction with specific antibody results in a color change. The simplicity of some of the tests allows them to be used in the clinic-office setting to which the majority of pandemic influenza-infected individuals with relatively mild illness are likely to present.
Although these POC kits were initially developed with seasonal influenza viruses in mind: QuickVue (42,149,151,184), BinaxNOW (15,42,51,62,114,154,169,175,184,206), Directigen Flu A+B and/or Directigen EZ Flu A+B (42,113,153,169,175,184,206), there was an immediate interest in testing them against the new pandemic virus. Unfortunately, it soon became clear that these rapid test kits appeared to be even less sensitive for the novel pandemic influenza virus, with sensitivities as low as 20%-30%, though most ranged over 40%-60% (14,29,56,63,67,77,86,105,110,123,164,165,193,198,199). Yet notably, in children who generally shed higher levels of virus, the reported sensitivities seemed to be generally higher (50,87,166), similar to seasonal influenza viruses (151,169), though there is some variability in this (199).
Therefore, for most of these rapid POC tests for both seasonal and the novel pandemic influenza virus, the overall sensitivity is poor, ranging from as low as 20% to as high as 90%, depending on which study is cited (86). Although they have been specifically designed to be easy to use (similar to an over-the-counter pregnancy test in complexity), their sensitivities vary according to many factors. These include the experience and skill of the operator, as well as other patient-related factors, such as age (they may be more sensitive in children who shed more virus), and the specific type of sample that is collected and tested. Although all types of respiratory samples can be used in such tests, the sensitivity appears to be better with nasopharyngeal swabs and aspirates than with throat swabs or gargles (48,163), which may contain more virus as it has been shown that the sensitivity of these rapid tests depend on the amount of virus in the sample (41,42,96,164,184). This in turn depends on how soon the patient presents for a sample to be collected after the onset of acute infection, as well as how well the sample (whether nasal swab, nasopharyngeal swab or nasopharyngeal aspirate) is taken (127).
In contrast, the specificity of these POC test kits is very good [specificity = no. of true negatives/ (no. of true negatives + false positives)], ranging over 70-100% for pandemic influenza (67,86,105,110), with much higher ranges (>90%) for seasonal influenza viruses (51,62,149,151,153,154,169,175,206). Again, some studies show some different outcomes for some of these kits, which may be due to the particular ‘gold standard assay’ against which they are tested (165), which is another variable that needs to be considered when comparing the performance data of these kits. A high specificity means that a positive result on the test is almost certainly an indication of influenza infection, rather than this just being a false positive. So for these rapid test kits, a positive result is more reliable (as an indicator of a true influenza infection) than a negative result (which may not necessarily indicate the absence of an influenza infection), in determining the patient’s influenza status.
Although many studies have been performed, comparing these rapid POC tests between themselves, as well as against the more conventional methods of viral culture, direct immunofluorescence and RT-PCR, it is difficult to say, definitively, which of these kits is the ‘best’ for diagnosing influenza infection in a patient. Thus, for a symptomatic patient with a negative result on one of these POC tests, this may not necessarily exclude an influenza infection and the physician needs to decide whether additional, more sensitive laboratory tests are required and likely to change their clinical management of the patient (including whether just to send the patient to the nearest emergency room). This will usually depend on the patient’s severity of clinical illness.
Antimicrobial Therapy
Antivirals
There are four antiviral drugs commonly used to treat influenza A and B. As well as the adamantane drugs (amantadine, rimantadine) that are only effective against influenza type A, there are the newer neuraminidase inhibitors (NAIs, zanamivir - Relenza®) and oseltamivir - Tamiflu®).
For seasonal influenza A(H3N2), avian influenza A(H5N1) and pandemic influenza A(H1N1), virtually all these viruses are resistant to treatment with the adamantane drugs (amatadine and rimantadine), but still susceptible to the neuraminidase inhibitors (NAIs - oseltamivir and zanamivir). For seasonal influenza A(H1N1), most of these viruses are resistant to oseltamivir (though zanamivir is still effective in most cases) and most are still susceptible to the adamantine drugs, though resistance seems to be increasing. Another member of the NAIs, peramivir, is still in clinical trials. This has an advantage over both oseltamivir (taken orally) and zanamivir (taken by inhaler) in that it can be given intravenously. Combination therapy with oseltamivir and rimantadine can be given empirically if the influenza subtype is unknown, and indeed some patients infected with 2009 A(H1N1) have been given this combination as initial empirical therapy (34).
According to the manufacturer’s information, oseltamivir is generally well tolerated and its adverse effects are mild, being mainly gastrointestinal (i.e. nausea, vomiting, diarrhea). However, reports from the use of oseltamivir as post-exposure prophylaxis (75 mg once daily for 10 days) in primary and secondary school children (age range 4-12 years) have described additional symptoms such as ‘feeling sick’, headaches, stomach aches, difficulties sleeping, nightmares and poor concentration (108,201).
Table 1 shows the currently recommended NAI doses for both treatment and post-exposure prophylaxis for different ages of patients. Treatment is recommended for 5 days, whereas prophylaxis is recommended for at least 2 weeks or at least for 7 days after contact and illness onset with the last infected individual. Pediatric dosing is based on weight for those weighing less than 40 kg, above 1 year of age. For children younger than 1 year, oseltamivir is not licensed, however it can be used on an ‘emergency authorization use’ basis, for which the recommended dosing regimen is also shown in Table 1. Note that careful monitoring for adverse effects should be conducted when used in this younger age group, outside the drug’s licensure.
The treatment of pregnant women infected with 2009 A(H1N1) is considered a priority as there appears to be an increased risk (though this is relative) of complications in this population (100). A recent study suggests that so far, oseltamivir treatment appears to be safe in pregnancy (179), and that on this basis, it should be commenced as soon as possible after the onset of symptoms (212), as well as being offered for post-exposure prophylaxis (35). However, as always, the actual application of these recommendations will be down to the individual decision and risk assessment of the patient and her doctor.
Antiviral Resistance
The mechanism of the adamantanes drugs (amantidine, rimantidine) is to block the viral M2 matrix protein, which is an ion channel required to cause a pH change to allow the uncoating of the virus and the release of its RNA, once it has been taken inside a permissive host cell by endocytosis (102,202). Several amino acid substitutions in this viral M2 gene will prevent the binding of these adamantine drugs to the M2 protein, thus conferring resistance to these drugs. These include: L26F (i.e. a change from leucine to phenylalanine at position 26 in the viral M2 gene), V27A (valine to alanine), A30T (alanine to threonine), S31N (serine to asparagine) and G34E (glycine to glutamic acid).
Similarly, drug resistance to the neuraminidase inhibitors (NAIs), oseltamivir and zanamivir, arise in the presence of amino acid mutations such as: H274Y (a switch from histidine to tyrosine) and N294S (asparagine to serine) in the N1 subtype of the viral neuraminidase, and R292K (arginine to lysine) and E119G/D/A/V (glutamic acid to glycine/ aspartic acid/ alanine/ valine) in the N2 subtype of the viral neuraminidase. Since all influenza B, most H3N2 seasonal influenza and the A(H1N1) pandemic influenza viruses are resistant to adamantanes (since they possess the M2 protein drug resistance mutations), it is recommended to use the neuraminidase inhibitors oseltamivir and zanamivir instead of amantadine and rimantadine for the treatment of current circulating influenza (54).
Resistance to oseltamivir arising in patients treated with oseltamivir has been reported in a few cases so far. The most commonly reported resistance mutation, H275Y (using the numbering for N2 viruses), occurs in the neuraminidase (NA) gene (36,37). The incidence and prevalence of oseltamivir-resistant 2009 A(H1N1) viruses is likely to increase, given the continued widespread use of the drug for treatment. However, now, oseltamivir should only be used for treating severely ill cases of 2009 A(H1N1) infection (as per seasonal influenza use recommendations) and no longer used as post-exposure prophylaxis unless vulnerable groups have been exposed (212). However, there has been one case of oseltamivir-resistance reported in an individual with no history of oseltamivir use (30). The worrying issue then is whether such resistant viruses will eventually become fit enough to transmit efficiently in the population (perhaps displacing the wild-type susceptible virus), making the worldwide stockpiles of oseltamivir effectively useless.
Antibiotics
There is increasing evidence and conviction that a significant proportion (20-30%) of deaths due past influenza pandemics may have been a result of secondary bacterial infections. Recent analyses of the 1918 pandemic mortality figures suggest that a significant number of deaths were due to secondary infection with Hemophilus influenzae, S. pneumoniae, S. pyogenes and/ or S. aureus (21). These findings suggest that both antibiotics and anti-bacterial vaccines may be an important component of the management of influenza infections (83).
Vaccines
Influenza immunization (or vaccination) should be the main method of controlling this infection and is recommended for certain high-risk groups in a population (Box 1) that may suffer more severe consequences of influenza infection, e.g. if they have pre-existing medical conditions that may predispose to worse clinical outcomes with the infection.
Each year the WHO Collaborating Laboratories from around the world gather a selection of influenza viruses isolated from clinical samples in order to identify which influenza subtypes and strains have been most recently circulating and which are most likely to persist in the population for some time. This allows them to recommend the most protective antigenic (almost entirely hemagglutinin, HA) composition for that season’s approaching influenza epidemics in the northern and southern hemispheres. Since these northern (Nov-Jan) and southern (Jun-Sep) influenza seasons are about 6 months apart, the vaccine manufacturers have, on average, just 6 months to produce enough vaccine for the northern and southern hemisphere populations, whilst still maintaining the high quality of the final product. Due to the variability of the viral genome, there are occasionally seasons where the vaccine antigen is not well matched to the circulating virus strain, and in these years, an excess morbidity and mortality can be seen (11,162) (Figure 6)
In recent years, the most common vaccine was an inactivated trivalent vaccine composed of the HA antigens from seasonal influenzas A(H1N1) and A(H3N2) and seasonal influenza B. However, since late 2009, the pandemic influenza A(H1N1) antigen (i.e. the A/California/7/2009 (H1N1)-like virus) has been incorporated instead of the seasonal A(H1N1) (i.e. A/Brisbane/59/2007-like virus) which appears to be disappearing from most regions (see Box 2).
Prior to the incorporation of the pandemic virus into the seasonal influenza vaccine, many trials were performed with the pandemic virus antigen alone as a monovalent vaccine with generally reasonable efficacy and relatively few adverse effects being reported (79,125,139,141,194), despite concomitant and intensified adverse effect surveillance and reporting (13,39,40). However, there have been more public adverse events reported in the world media, including some incidences of febrile seizures (see Table 2), occasional severe injection site lymphadenitis (82,169), and perhaps most unusual of all, narcolepsy in vaccine recipients in Sweden (132). Investigations into some of these adverse effects are being conducted, but in the vast majority of individuals, the inactivated trivalent seasonal influenza vaccine containing the 2009 pandemic influenza A(H1N1) virus (of swine origin) has so far been found to be just as safe as the previous seasonal influenza vaccines (196).
Such inactivated vaccines can take a variety of forms in the way the virus antigen is prepared, including inactivated whole or split virus or subunit virus antigens. In recent years, there has been heightened concern regarding other components of vaccines, including the various types of manufacturers’ proprietary adjuvants and in particular, the mercury-containing preservative, thiomersal.
Thiomersal has now been removed from most vaccines routinely recommended for children of 6 years or younger, though there are still some influenza vaccines that contain thiomersal, some of which are shown in the Table 2 below with their specific mercury content listed. The main risk is that of an allergic reaction in the form of a local delayed-type hypersensitivity reaction. However, very few people exhibit any such reaction in response to the vaccine – even those who display such a reaction to thiomersal in patch testing.
Live attenuated vaccines have never contained thiomersal and one such vaccine is available for influenza, though the age range for which these are licensed are more restrictive than for the more traditional inactivated vaccines. In addition, this vaccine is contraindicated in young children (aged 2-4 years) with or at risk of asthma (e.g. those with a history of wheezing within the last 12 months). More information on the use of thiomersal in vaccines is available at the US Food and Drug Administration (FDA) website (69).
There has been interest in recent years to move towards developing a more universal influenza vaccine (i.e. without the need to update and manufacture new vaccine batched each year), which contains more conserved influenza virus antigens that remain relatively similar across the many different virus subtypes (i.e. H1, H3, H5, H7, H9, etc.), but these vaccines are still very much experimental (57,68,167,176), and the current WHO-led surveillance and updating of the antigenic components of the annual seasonal influenza vaccines for northern and southern hemispheres may well continue for many years to come.
Prevention
Public Health Issues
Even before the emergence of the 2009 influenza A(H1N1) pandemic, in recent years there has been a greater emphasis on public health interventions to inhibit human-to-human transmission of influenza, which has led to a renewed interest in the study of the epidemiology and transmission of these viruses (78,97,117,133,189). Reinforcing this, there have been a number of public health threats related to various subtypes of animal influenza viruses infecting humans in recent years (e.g. H5, H7, H9, as reviewed earlier in the Introduction). One of these, avian influenza H5N1, had already raised concerns about the potential to cause a pandemic and many of the existing pandemic preparedness plans had been based on this threat from H5N1 (119,128,215). Hence, it was something of a surprise to many agencies, worldwide, when the novel swine-origin influenza A (H1N1) virus (S-OIV) emerged from Mexico leading to the WHO, in June 2009, declaring that its spread had reached pandemic proportions (43).
After its emergence from Mexico in March 2009, it was soon found that because of its novelty and therefore the absence of any prior immunity in most of the population, the 2009 pandemic influenza A(H1N1) virus transmitted efficiently between individuals even outside the normal seasonal influenza period for a particular country. Early estimates put the basic reproduction number (R0 – the expected number of secondary cases when exposed to a single index case in a fully susceptible population) at between 1.4–1.6 and 2.2–3.1, using different analyses of the Mexican 2009 A(H1N1) data (31). Secondary attack rates in the well-defined outbreak reported from the Mexican town of La Gloria, Veracruz, in mid-February of 2009 were estimated at 61% and 29% in those under and over the age of 15 years, respectively (70).
One of the first steps for public health teams in identifying and controlling the spread of the disease is to provide an accurate, yet easy-to-apply case definition. Case definitions form the cornerstone of the investigation and management of outbreaks – duties that form a significant component of the role of public health agencies. Early on in the 2009 influenza A(H1N1) pandemic, the WHO developed case definitions for cases of this new infection (Box 3). Interestingly, they also included laboratory test results in these definitions, which was unusual – most case definitions are made purely on clinical symptoms and signs (see Box 3).
However, purely clinical case definitions may not be specific enough and include many cases that are unrelated to the specific pathogen under investigation. One interesting study evaluated the accuracy of clinical case definitions for different influenza subtypes during an outbreak in France during the 1995–1996 influenza season. Predictors of influenza virus infection were selected with logistic regression models. The results varied with the influenza virus subtype: temperature of >38.2oC, stiffness or myalgia, rhinorrhea, and cough were more predictive of influenza A(H3N2), whereas fatigue, lacrimation or conjunctival injection, and the absence of stiffness or myalgia were more predictive of influenza A(H1N1). In all 12 clinical case definitions were evaluated for their abilities to diagnose influenza virus infection. They were associated with positive predictive values of 27% to 40% and negative predictive values of 80% to 91% (24).
In a situation such as the current 2009 swine-origin influenza A(H1N1) pandemic, where most countries affected have abandoned testing (simply because the number of samples is far too high) and rely on clinical case definitions, a comparative evaluation of case definitions of influenza in subsets of patients would help inform optimal estimates of the disease burden associated with influenza epidemics. However, in most outbreak situations, only one consensus case definition will be used and such retrospective analyses of its effectiveness can only be performed later – usually once the outbreak is well underway, or even when it is over.
Infection Control Measures
Whereas public health tends to focus on limiting disease impact at a population level, infection control focuses more on limiting the transmission of infection between individuals, e.g. between family members, doctors and their patients, etc. Unfortunately, a study of the literature to date indicates that the quality of evidence upon which disease mitigation can be planned is generally poor (9,10). Aledort et al. evaluated the relative merits of non-pharmaceutical interventions by combining available evidence from the literature with qualitative and quantitative expert opinion (2). Non-pharmaceutical public health interventions that could potentially mitigate an influenza pandemic were grouped into four categories (summarized in Table 3):
(1) infection control and prevention to reduce transmission when contact occurs between infected and uninfected individuals;
(2) patient management to reduce contact between infected and uninfected individuals;
(3) contact management to reduce contact between possibly infected and uninfected individuals;
(4) voluntary and mandatory community restrictions to reduce contact between groups that may contain infected persons.
Amongst the most common interventions recommended the following deserve a more detailed mention:
Hand Hygiene
Infection control doctrines such as hand hygiene and respiratory etiquette are widely supported in the literature and universally accepted (101,148). The WHO's recommended pandemic influenza interventions include hand hygiene as routine (10). Other controlled studies have shown a protective effect of hand hygiene in reducing upper respiratory infections, although not all of these are necessarily related to influenza infection (64,173). The recommendation therefore is that rigorous and routine hand hygiene using either soap and water or a suitable hand gel sanitizer is an important strategy for healthcare workers and the general public in all settings and at all phases of a pandemic (2).
Respiratory Etiquette
Respiratory etiquette is another important means of preventing transmission in all pandemic phases. Respiratory hygiene and cough etiquette is generally interpreted to include covering the mouth and nose with a tissue or into the upper sleeve when coughing or sneezing, and refraining from spitting (38). Promotion of respiratory etiquette should be coupled with a strong emphasis on public education that includes all the relevant information about influenza.
Human Surveillance and Case Reporting
Whilst there is no direct evidence on the usefulness of surveillance and case reporting in the context of influenza transmission, experts agree that influenza surveillance supports a range of necessary preparedness activities. There are concerns, however, about resource constraints, especially in a large outbreak (2).
Rapid Viral Diagnosis
In most countries viral diagnosis of influenza is currently not routine practice, and is hampered by lack of resources, the need for expert technologists and expensive equipment. Novel point of care sensitive tests, need to be available in a way that facilitates its use in non-clinical settings especially if they are to be useful in a large pandemic situation, but again, performance (mainly test sensitivity) and cost is the main issue as most of these ‘point-of-care’ test kits are relatively expensive. A selection of these rapid test kits is reviewed under the Diagnosis section.
Use of Masks and Other Personal Protective Equipment (PPE)
Droplet transmission is thought to be the primary mode of transmission of the influenza virus. The CDC guidelines therefore suggest that health-care personnel wear masks only for close patient contact (i.e., within 3 feet) (32).
There is limited evidence from seasonal influenza to support and differentiate between the clinical importance of contact, droplet or droplet nuclei (airborne) transmission of influenza. A belief in the potential risk of airborne transmission has lent support for N95 respirators, which are designed to stop up to 95% of small airborne particles (18). This area is controversial and the evidence is limited (17,116,171,180,185,186,187), though more recent studies have certainly demonstrated that both influenza RNA and live virus can be found in air (12,120,177,218), so ignoring this potential route of airborne transmission may now be imprudent. Interestingly, some individuals have taken such risks seriously enough to start developing their own interventions to ‘catch’ coughs and sneezes in an effort to limit transmission of infection to others [for example, see: http://www.coughcatcher.com/].
Current thinking recommends reserving surgical masks, N95 respirators and other personal protective equipment for hospital and ambulatory patients when the pandemic is widespread. It is noteworthy that N95 respirators need fit-testing and are expensive, further compromising their usefulness. Discomfort is also a problem when N95 masks have to be worn for long periods and several studies of compliance with N95 masks have found a general reluctance amongst healthcare workers to comply with these recommendations – even in potentially hazardous situations (74,130,168).
Interestingly, a recent study has shown that surgical and N95 masks may provide the same level of protection in healthcare workers, using the detection of influenza infection in participants by PCR or 4-fold rise in serological titers as the endpoint for the study (122). In this study of 478 nurses randomly assigned to receive either surgical masks or N95 respirators, influenza infection occurred in 50 nurses (23.6%) in the surgical mask group and in 48 (22.9%)in the N95 respirator group (absolute risk difference, −0.73%; 95% CI, −8.8% to 7.3%;P=.86), the lower confidence limit being inside the non-inferiority limit of −9%. This single study suggested that use of surgical masks compared with an N95 respirator resulted in non-inferior rates of laboratory confirmed influenza (122). However, there are many potential confounders in this study (some of which are acknowledged by the authors themselves) and other teams should repeat this study before such counter-intuitive conclusions about the comparative efficiencies of surgical and N95 masks are widely accepted.
In contrast, the use of masks and other personal protective equipment for the general public has even less evidence to support it with the exception of some evidence from the SARS outbreak, which may not be relevant as the behaviour of the SARS-coronavirus and influenza are quite different, i.e. SARS was shown to be relatively uninfectious until the appearance of symptoms, whereas influenza does appear to have a short period of transmissibility prior to symptom onset (71). Most experts recommend against the use of masks or respirators by the public prior to the arrival of pandemic influenza and in the early localized phase (2).
Isolation of the Sick
The evidence for mandatory isolation in any specific setting is inconclusive. The concern is that healthcare facilities are likely to be rapidly overwhelmed. However, more recent studies report that when numbers are small, isolation in hospitals using appropriate infection control measures may be effective (8). In spite of the lack of definitive evidence, voluntary self-isolation in the home was recommended for all phases of a US pandemic (2). An additional factor to take into account is the lethality of the infection that is being controlled. For a highly lethal and transmissible pathogen, such as smallpox, isolation of the sick and quarantining of those exposed is more acceptable to the general public and therefore probably more effective, as the consequences of not limiting the spread throughout the population are more devastating.
Social Distancing
Social distancing measures such as office and school closures, cancellation of large gatherings at events, and travel restrictions have been implemented in many countries during SARS and more recently during the current swine-origin influenza A (H1N1) pandemic. However, their effectiveness in an influenza outbreak has not yet been well-established (10,65). Despite the current suggestion that influenza epidemics may be amplified in primary schools, data on the effectiveness of school closures for reducing community transmission are contradictory. Some studies suggest a decline in community transmission rates of respiratory infections with school closures (90), whereas others have noted a protective effect when schools remained open (10).
Recent modeling studies generally support school closure and confinement in the home as an effective means of reducing overall attack rates within communities when coupled with antiviral prophylaxis, however the reality remains that this does not account for the effect that this may have when individuals are displaced into other settings (8,73), though one recent questionnaire-based study has demonstrated the overall number of contacts does decrease for individual school children when schools are closed (98).
One well-investigated example of this has been the effects of school closure on the subsequent progression of an influenza pandemic. Various mathematical modeling studies have suggested that whilst there maybe some delay in the spread of the pandemic from closing schools, this will only be effective if the subsequent behavior of children outside of school does not result in a similar number of contacts and that such relative isolation is prolonged during the pandemic period (107). In addition, the social and economic disruption for the working parents may be difficult to overcome as at least one parent will have to take time off work to look after young children at home (26,27). In particular, school closure has to be part of an overall mitigation strategy, including the treatment and home isolation of infected individuals to reduce further contacts (178).
Screening Travelers and Travel Restrictions
Consistent with the literature, widespread and sustained screening of travelers would ultimately be impractical and inefficient as long as detecting asymptomatic shedding is not feasible. Screening and quarantining entering travelers at international borders did not substantially delay virus introduction in past pandemics, except in some island countries, and will likely be even less effective in the modern era. Instead, WHO recommends providing information to international travelers and possibly screening travelers departing countries with potentially transmissible human infections. The principal focus of interventions against pandemic influenza spread should be at national and community levels rather than international borders (9). Various mathematical modeling tools have been used to assess the impact of restricting air travel on the spread of pandemic influenza. However, the benefits may be fairly minor and may not be worth the inevitable and serious social and economic disruption this will cause (20,61,91).
Many teams, worldwide are working on the better understanding of various aspects of the way influenza transmits in different situations and as our knowledge improves it is likely that these public health approaches will be modified.
Conclusions
The continued variation and adaptation of the influenza virus to different hosts (both animal and human) in different environments will continue to challenge us. Ongoing surveillance of influenza viral antigenic drift for the annual update of the antigenic composition of the seasonal influenza vaccine constitutes one of the most successful international public health collaborations in modern times. However, the threat from influenza is two-tiered. As was seen with the recent 2009 influenza A(H1N1) pandemic, the surveillance for such influenza viral reassortment or antigenic shift events is much more incomplete as it needs to involve various animal species that can also act as influenza virus reservoirs (145,200). Such coverage is much more problematic as the numbers of such species, any one of which could be harbouring a unique reassortment, is vast and scattered in varying livestock holdings across the globe. This will require much closer and regular cooperation with the farming and veterinary community in order to comprehensively characterize and monitor these animal viral populations. This will only become more essential as the world population increases and the demand for high quality meat, requiring more intensive farming methods of such animal hosts, only increases. However, with sufficient support and funding from governments as well as those in the livestock industry, together with the ongoing dedication of the clinical and scientific influenza virus community, there is no reason why the level of animal influenza virus surveillance should not be able to at least match that which already exists for human influenza.
CLINICAL CASE SECTION
Case A
Patient A was a 41-year old Caucasian man with no underlying comorbidities and no history of influenza vaccination. He was admitted to hospital with a 4-day history of a flu-like illness, including rhinorrhea, nausea, vomiting and diarrhea. He had been initially treated with oral empirical antibiotics (amoxicillin and clarithromycin – 500 mg tds and 500 mg bd respectively) by his general practitioner (GP) at the onset of his illness, which was later converted to IV cefuroxime (1.5 g tds) and clarithromycin (500 mg bd oral).On admission, he was thrombocytopenic (63 x109 /L), neutropenic (0.80x109 /L) and lymphopenic (0.60 x109 /L), with an overall leukopenia (total white cell count of 1.44 x109 /L) His alanine aminotransferase (ALT) was also elevated at 132 iu/L. His highest recorded temperature was 37.5oC and his C-reactive protein (CRP) peaked at 234.8 mg/L on Day 4 of his admission. He was started on oseltamivir (75 mg oral twice daily), empirically, on admission. Pandemic influenza A(H1N1) infection was confirmed by PCR testing on throat and nasal swabs the next day, by which time he had been switched to intravenous (IV) zanamivir as he was unable to absorb orally. Despite this, he rapidly deteriorated (see chest X-ray series below) and was admitted to intensive care (ICU) the next day, where he eventually died 6 days later. No evidence of secondary bacterial infection was found. There was no evidence of hemorrhagic viral pneumonia. No testing was performed for the detection of the viral oseltamivir-resistance mutation H275Y. No other respiratory virus out of: influenza A (non-H1N1 pandemic) and B, respiratory syncytial virus (RSV), parainfluenza viruses, human metapneumovirus or adenoviruses were detected. His family, a wife and two young children (twins) were also infected with pandemic A(H1N1) but recovered without any serious illness. In summary, this was a rapidly fatal case of pandemic A(H1N1) infection in a previously healthy man with no underlying comorbidities. Cytokine studies were not performed, though may have proved interesting.
Figure A: Serial chest X-ray changes for Patient A in a fatal pandemic influenza A(H1N1) infection.
Case B
Patient B was a 22-year old, pregnant (29 weeks gestation) Somalian woman with no underlying comorbidities and no history of influenza vaccination. She was seen after a 5-day history of flu-like illness (coryzal symptoms, blocked nose) which had worsened over the previous day with fever and dyspnea. Other members of her family had also developed a flu-like illness. She was admitted (Day 1) under the obstetric team and treated for community-acquired pneumonia (CAP) with oral amoxicillin 500 mg tds and clarithromycin 500 mg bd. Her highest recorded temperature was 38oC. Her CRP was elevated at 133.2 mg/L on admission and her alkaline phosphatase was raised at 196 iu/L. She began to deteriorate overnight and was given non-invasive ventilation whilst awaiting transfer to ICU, where she was eventually intubated for worsening respiratory function and pulmonary edema. The following day (Day 2), pandemic influenza A(H1N1) was detected in a respiratory sample and she was started on oseltamivir (dose 75 mg bd orally). No other respiratory virus out of: seasonal influenza A(H1N1) and B, respiratory syncytial virus (RSV), parainfluenza viruses, human metapneumonvirus or adenoviruses were detected. No other coinfecting bacteria were found at this point, but she had been on empirical antibiotics. On ICU during the same day, she desaturated to 47% during suctioning and large amounts of reddish fluid (possibly a sign of a viral hemorrhagic pneumonitis) was removed. As she was obviously not ventilating adequately a decision was made to deliver the baby via emergency Caesarian section the same day. The baby was found to be negative for pandemic influenza A(H1N1) and was stable on neonatal ICU on continuous positive airways pressure (CPAP) ventilation. Soon after, the mother had to be transferred to another hospital for respiratory monitoring for possible extracorporeal membrane oxygenation (ECMO). She was found to be improving after several days, and was eventually transferred back to the original hospital without undergoing ECMO (Day 9). Her CRP had risen to a high of 321 mg/L on Day 10 before decreasing thereafter. She continued to improve on ICU and was extubated on day 13 and transferred to the obstetric unit. Her treatment with oseltamivir ceased on Day 12 as two repeat pandemic influenza A(H1N1) PCRs were found to be negative. She was eventually discharged home, fully recovered with no further complications.
Case C
Patient C was a 45-year old Caucasian man with no underlying comorbidities and no history of influenza vaccination. He was admitted to two other hospitals prior to the current admission. His initial presentation to the first hospital (duration of stay 5 days) was one of severe respiratory failure and septicemia after a week of flu-like symptoms. Pandemic influenza A(H1N1) was detected and treatment was started with oseltamivir (75 mg bd orally). His blood cultures grew a Group A Streptococcus, for which he was he was treated with intravenous benzylpenicillin 1.2 g qds. His subsequent sepsis rapidly led to renal failure, requiring filtration. His respiratory failure required extracorporeal membrane oxygenation (ECMO) at the second hospital (duration of stay approximately 3 weeks). He was eventually transferred to the current hospital (Day 1 of admission) after coming off ECMO but still requiring ventilatory support on ICU and hemofiltration. On admission his C-reactive protein was markedly elevated at 195.3 mg/L, with anemia (Hb 9.8 g/dL), lymphopenia (0.50 x109 /L) and a mild neutrophilia (9.16 x109 /L). His renal function was still poor with a urea of 39.0 mmol/L and a creatinine of 409 mmol/L. He also a mild obstructive liver profile with a raised alkaline phosphatase at 135 iu/L and an elevated gamma glutamyl transferase level of 206 iu/L. Treatment with oseltamivir continued and on Day 3 his pandemic influenza A(H1N1) PCR was still positive. A tracheostomy was performed and he received support through a tracheostomy mask. His A(H1N1) PCR became negative by Day 8. Interestingly, a repeat check showed a recurrence of a low level A(H1N1) positive PCR on Day 9. In view of this persistent infection, oseltamivir was continued. Eventually, he became PCR negative on two separate samples by Day 22 and 23, and was stable enough to have his tracheostomy decannulated and to move to a hospital ward by Day 23. He was eventually discharged home, fully recovered with no further complications.
References
1. Agyeman P, Duppenthaler A, Heininger U, Aebi C. Influenza-associated myositis in children. Infection 2004;32:199-203. [PubMed]
2. Aledort JE, Lurie N, Wasserman J, Bozzette SA. Non-pharmaceutical public health interventions for pandemic influenza: an evaluation of the evidence base. BMC Public Health 2007;7:208. [PubMed]
3. Baltagi SA, Shoykhet M, Felmet K, Kochanek PM, Bell MJ. Neurological sequelae of 2009 influenza A (H1N1) in children: a case series observed during a pandemic. Pediatr Crit Care Med 2010;11:179-184. [PubMed]
4. Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, Garcia-Sastre A, Palese P, Taubenberger JK. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci U S A 2001;98:2746-2751. [PubMed]
5. Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, Zaki SR, Hayden FG, Hui DS, Kettner JD, Kumar A, Lim M, Shindo N, Penn C, Nicholson KG. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med 2010;362:1708-1719. [PubMed]
6. Bay A, Etlik O, Oner AF, Unal O, Arslan H, Bora A, Davran R, Yuca SA, Dogan M. Radiological and clinical course of pneumonia in patients with avian influenza H5N1. Eur J Radiol 2007;61:245-250. [PubMed]
7. Bean WJ, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster RG. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J Virol 1992;66:1129-1138. [PubMed]
8. Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. Avian influenza A (H5N1) infection in humans. N Engl J Med 2005;353:1374-1385. [PubMed]
9. Bell DM. Non-pharmaceutical interventions for pandemic influenza, international measures. Emerg Infect Dis 2006;12:81-87.[PubMed]
10. Bell DM. Non-pharmaceutical interventions for pandemic influenza, national and community measures. Emerg Infect Dis 2006;12:88-94. [PubMed]
11. Bhat N, Wright JG, Broder KR, Murray EL, Greenberg ME, Glover MJ, Likos AM, Posey DL, Klimov A, Lindstrom SE, Balish A, Medina MJ, Wallis TR, Guarner J, Paddock CD, Shieh WJ, Zaki SR, Sejvar JJ, Shay DK, Harper SA, Cox NJ, Fukuda K, Uyeki TM. Influenza-associated deaths among children in the United States, 2003-2004. N Engl J Med 2005;353:2559-2567. [PubMed]
12. Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, Meade BJ, Lander O, Davis S, Thewlis RE, Celik I, Chen BT, Beezhold DH. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis 2009;48:438-440. [PubMed]
13. Black S, Eskola J, Siegrist CA, Halsey N, Macdonald N, Law B, Miller E, Andrews N, Stowe J, Salmon D, Vannice K, Izurieta HS, Akhtar A, Gold M, Oselka G, Zuber P, Pfeifer D, Vellozzi C. Importance of background rates of disease in assessment of vaccine safety during mass immunisation with pandemic H1N1 influenza vaccines. Lancet 2009;374:2115-2122. [PubMed]
14. Blyth CC, Iredell JR, Dwyer DE. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;361:2493. [PubMed]
15. Booth S, Baleriola C, Rawlinson WD. Comparison of two rapid influenza A/B test kits with reference methods showing high specificity and sensitivity for influenza A infection. J Med Virol 2006;78:619-622. [PubMed]
16. Bragstad K, Jorgensen PH, Handberg K, Hammer AS, Kabell S, Fomsgaard A. First introduction of highly pathogenic H5N1 avian influenza A viruses in wild and domestic birds in Denmark, Northern Europe. Virol J 2007;4:43. [PubMed]
17. Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Transmission of influenza A in human beings. Lancet Infect Dis 2007;7:257-265. [PubMed]
18. Bridges CB, Kuehnert MJ, Hall CB. Transmission of influenza: implications for control in health care settings. Clin Infect Dis 2003;37:1094-1101. [PubMed]
19. Brown CM. Severe influenza A virus (H1N1) infection in pregnancy. Obstet Gynecol 2010;115:412-414. [PubMed]
20. Brownstein JS, Wolfe CJ, Mandl KD. Empirical evidence for the effect of airline travel on inter-regional influenza spread in the United States. PLoS Med 2006;3:e401. [PubMed]
21. Brundage JF, Shanks GD. Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg Infect Dis 2008;14:1193-1199. [PubMed]
22. Butt AM, Siddique S, Idrees M, Tong Y. Avian influenza A (H9N2): computational molecular analysis and phylogenetic characterization of viral surface proteins isolated between 1997 and 2009 from the human population. Virol J 2010;7:319. [PubMed]
23. Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 2005;43:5760-5767. [PubMed]
24. Carrat F, Tachet A, Rouzioux C, Housset B, Valleron AJ. Evaluation of clinical case definitions of influenza: detailed investigation of patients during the 1995-1996 epidemic in France. Clin Infect Dis 1999;28:283-290. [PubMed]
25. Cauchemez S, Donnelly CA, Reed C, Ghani AC, Fraser C, Kent CK, Finelli L, Ferguson NM. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med 2009;361:2619-2627. [PubMed]
26. Cauchemez S, Ferguson NM, Wachtel C, Tegnell A, Saour G, Duncan B, Nicoll A. Closure of schools during an influenza pandemic. Lancet Infect Dis 2009;9:473-481. [PubMed]
27. Cauchemez S, Valleron AJ, Boelle PY, Flahault A, Ferguson NM. Estimating the impact of school closure on influenza transmission from Sentinel data. Nature 2008;452:750-754. [PubMed]
28. Centers for Disease Control and Prevention (CDC). Bacterial Coinfections in Lung Tissue Specimens from Fatal Cases of 2009 Pandemic Influenza A (H1N1) - United States,May - August 2009. MMWR Morb Mortal Wkly Rep 2009:1071-1074. [PubMed]
29. Centers for Disease Control and Prevention (CDC). Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus - United States, 2009. MMWR Morb Mortal Wkly Rep 2009;58:826-829. [PubMed]
30. Centers for Disease Control and Prevention (CDC). Hong Kong oseltamivir resistance. Available at:http://www.cdc.gov/h1n1flu/HAN/070909.htm. Accessed on August 28, 2009.
31. Centers for Disease Control and Prevention (CDC). Human infection with new influenza A (H1N1) virus: clinical observations from Mexico and other affected countries, May 2009. . Wkly Epidemiol Rec 2009; 84:185-189 2009. [PubMed]
32. Centers for Disease Control and Prevention (CDC). Interim Guidance for the Use of Masks to Control Influenza Transmission. Available at: http://www.cdc.gov/flu/professionals/infectioncontrol/maskguidance.htm. Accessed on February 25, 2011.
33. Centers for Disease Control and Prevention (CDC). Interim Recommendations for Clinical Use of Influenza Diagnostic Tests During the 2009-10 Influenza Season. Available at: http://www.cdc.gov/h1n1flu/guidance/diagnostic_tests.htm. Accessed on 26 January 2011.
34. Centers for Disease Control and Prevention (CDC). Neurologic complications associated with novel influenza A (H1N1) virus infection in children - Dallas, Texas, May 2009. MMWR Morb Mortal Wkly Rep 2009;58:773-778. [PubMed]
35. Centers for Disease Control and Prevention (CDC). Novel influenza A (H1N1) virus infections in three pregnant women - United States, April-May 2009. MMWR Morb Mortal Wkly Rep 2009;58:497-500. [PubMed]
36. Centers for Disease Control and Prevention (CDC). Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis--North Carolina, 2009. MMWR Morb Mortal Wkly Rep 2009;58:969-972. [PubMed]
37. Centers for Disease Control and Prevention (CDC). Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients - Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep 2009;58:893-896. [PubMed]
38. Centers for Disease Control and Prevention (CDC). Respiratory Hygiene/Cough Etiquette in Healthcare Settings. Available at:http://www.cdc.gov/flu/professionals/infectioncontrol/resphygiene.htm. Accessed on 26 January, 2011.
39. Centers for Disease Control and Prevention (CDC). Safety of influenza A (H1N1) 2009 monovalent vaccines - United States, October 1-November 24, 2009. MMWR Morb Mortal Wkly Rep 2009;58:1351-1356. [PubMed]
40. Centers for Disease Control and Prevention (CDC). Preliminary results: surveillance for Guillain-Barre syndrome after receipt of influenza A (H1N1) 2009 monovalent vaccine - United States, 2009-2010. MMWR Morb Mortal Wkly Rep 2010;59:657-661.[PubMed]
41. Chan KH, Lai ST, Poon LL, Guan Y, Yuen KY, Peiris JS. Analytical sensitivity of rapid influenza antigen detection tests for swine-origin influenza virus (H1N1). J Clin Virol 2009;45:205-207. [PubMed]
42. Chan KH, Lam SY, Puthavathana P, Nguyen TD, Long HT, Pang CM, Chan KM, Cheung CY, Seto WH, Peiris JS. Comparative analytical sensitivities of six rapid influenza A antigen detection test kits for detection of influenza A subtypes H1N1, H3N2 and H5N1. J Clin Virol 2007;38:169-171. [PubMed]
43. Chan M. World now at the start of 2009 influenza pandemic. Available at:http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/print.html Accessed on February 25, 2011.
44. Chan PK. Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis 2002;34 Suppl 2:S58-64.[PubMed]
45. Chang LY, Lee PI, Lin YJ, Chiu HH, Huang LM, Lee CY. Influenza B virus infection associated with shock in a two-month-old infant. J Formos Med Assoc 1996;95:703-705. [PubMed]
46. Chotpitayasunondh T, Ungchusak K, Hanshaoworakul W, Chunsuthiwat S, Sawanpanyalert P, Kijphati R, Lochindarat S, Srisan P, Suwan P, Osotthanakorn Y, Anantasetagoon T, Kanjanawasri S, Tanupattarachai S, Weerakul J, Chaiwirattana R, Maneerattanaporn M, Poolsavathitikool R, Chokephaibulkit K, Apisarnthanarak A, Dowell SF. Human disease from influenza A (H5N1), Thailand, 2004. Emerg Infect Dis 2005;11:201-209. [PubMed]
47. Cohen D, Carter P. Conflicts of interest. WHO and the pandemic flu "conspiracies". BMJ 2010;340:c2912. [PubMed]
48. Covalciuc KA, Webb KH, Carlson CA. Comparison of four clinical specimen types for detection of influenza A and B viruses by optical immunoassay (FLU OIA test) and cell culture methods. J Clin Microbiol 1999;37:3971-3974. [PubMed]
49. Cox NJ, Subbarao K. Influenza. Lancet 1999;354:1277-1282. [PubMed]
50. Cruz AT, Demmler-Harrison GJ, Caviness AC, Buffone GJ, Revell PA. Performance of a rapid influenza test in children during the H1N1 2009 influenza a outbreak. Pediatrics 2010;125:e645-650. [PubMed]
51. Dale SE, Mayer C, Mayer MC, Menegus MA. Analytical and clinical sensitivity of the 3M rapid detection influenza A+B assay. J Clin Microbiol 2008;46:3804-3807. [PubMed]
52. Davies A, Jones D, Bailey M, Beca J, Bellomo R, Blackwell N, Forrest P, Gattas D, Granger E, Herkes R, Jackson A, McGuinness S, Nair P, Pellegrino V, Pettila V, Plunkett B, Pye R, Torzillo P, Webb S, Wilson M, Ziegenfuss M. Extracorporeal Membrane Oxygenation for 2009 Influenza A(H1N1) Acute Respiratory Distress Syndrome. JAMA 2009;302:1888-1895. [PubMed]
53. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;360:2605-2615. [PubMed]
54. Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis 2007;196:249-257. [PubMed]
55. Donaldson LJ, Rutter PD, Ellis BM, Greaves FE, Mytton OT, Pebody RG, Yardley IE. Mortality from pandemic A/H1N1 2009 influenza in England: public health surveillance study. BMJ 2009;339:b5213. [PubMed]
56. Drexler JF, Helmer A, Kirberg H, Reber U, Panning M, Muller M, Hofling K, Matz B, Drosten C, Eis-Hubinger AM. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis 2009;15:1662-1664. [PubMed]
57. Du L, Zhou Y, Jiang S. Research and development of universal influenza vaccines. Microbes Infect 2010;12:280-286. [PubMed]
58. Du Ry van Beest Holle M, Meijer A, Koopmans M, de Jager CM. Human-to-human transmission of avian influenza A/H7N7, The Netherlands, 2003. Euro Surveill 2005;10:264-268. [PubMed]
59. Ducatez MF, Olinger CM, Owoade AA, De Landtsheer S, Ammerlaan W, Niesters HG, Osterhaus AD, Fouchier RA, Muller CP. Avian flu: multiple introductions of H5N1 in Nigeria. Nature 2006;442:37. [PubMed]
60. Ekstrand JJ, Herbener A, Rawlings J, Turney B, Ampofo K, Korgenski EK, Bonkowsky JL. Heightened neurologic complications in children with pandemic H1N1 influenza. Ann Neurol 2010;68:762-766. [PubMed]
61. Epstein JM, Goedecke DM, Yu F, Morris RJ, Wagener DK, Bobashev GV. Controlling pandemic flu: the value of international air travel restrictions. PLoS One 2007;2:e401. [PubMed]
62. Fader RC. Comparison of the Binax NOW Flu A enzyme immunochromatographic assay and R-Mix shell vial culture for the 2003-2004 influenza season. J Clin Microbiol 2005;43:6133-6135. [PubMed]
63. Faix DJ, Sherman SS, Waterman SH. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009;361:728-729. [PubMed]
64. Falsey AR, Criddle MM, Kolassa JE, McCann RM, Brower CA, Hall WJ. Evaluation of a handwashing intervention to reduce respiratory illness rates in senior day-care centers. Infect Control Hosp Epidemiol 1999;20:200-202. [PubMed]
65. Ferguson NM, Cummings DA, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, Burke DS. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 2005;437:209-214. [PubMed]
66. Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature 2003;422:428-433. [PubMed]
67. Fernandez C, Cataletto M, Lee P, Feuerman M, Krilov L. Rapid influenza A testing for novel H1N1: point-of-care performance. Postgrad Med 2010;122:28-33. [PubMed]
68. Fiers W, De Filette M, El Bakkouri K, Schepens B, Roose K, Schotsaert M, Birkett A, Saelens X. M2e-based universal influenza A vaccine. Vaccine 2009;27:6280-6283. [PubMed]
69. Food and Drug Administration (FDA). Thimerosal in Vaccines. Available at:http://www.fda.gov/BiologicsBloodVaccines/SafetyAvailability/VaccineSafety/UCM096228. Accessed on February 25, 2011.
70. Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez-Gatell H, Alpuche-Aranda CM, Chapela IB, Zavala EP, Guevara DM, Checchi F, Garcia E, Hugonnet S, Roth C. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009;324:1557-1561. [PubMed]
71. Fraser C, Riley S, Anderson RM, Ferguson NM. Factors that make an infectious disease outbreak controllable. Proc Natl Acad Sci U S A 2004;101:6146-6151. [PubMed]
72. Gaydos JC, Top FH, Jr., Hodder RA, Russell PK. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 2006;12:23-28. [PubMed]
73. Germann TC, Kadau K, Longini IM, Jr., Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 2006;103:5935-5940. [PubMed]
74. Gershon RR, Pearson JM, Westra LJ. Evaluation tool for the assessment of personal protective respiratory equipment. Infect Control Hosp Epidemiol 2009;30:716-718. [PubMed]
75. Gibbs MJ, Armstrong JS, Gibbs AJ. Recombination in the hemagglutinin gene of the 1918 "Spanish flu". Science 2001;293:1842-1845. [PubMed]
76. Gibbs MJ, Gibbs AJ. Molecular virology: was the 1918 pandemic caused by a bird flu? Nature 2006;440:E8; discussion E9-10.[PubMed]
77. Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, Falk L, Lotlikar M, Kowerska M, Becker G, Korologos D, de Geronimo M, Crawford JM. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol 2009;45:191-195. [PubMed]
78. Gostin L. Public health strategies for pandemic influenza: ethics and the law. JAMA 2006;295:1700-1704. [PubMed]
79. Greenberg ME, Lai MH, Hartel GF, Wichems CH, Gittleson C, Bennet J, Dawson G, Hu W, Leggio C, Washington D, Basser RL. Response to a monovalent 2009 influenza A (H1N1) vaccine. N Engl J Med 2009;361:2405-2413. [PubMed]
80. Gregg MB, Hinman AR, Craven RB. The Russian flu. Its history and implications for this year's influenza season. Jama 1978;240:2260-2263. [PubMed]
81. Gregory V, Lim W, Cameron K, Bennett M, Marozin S, Klimov A, Hall H, Cox N, Hay A, Lin YP. Infection of a child in Hong Kong by an influenza A H3N2 virus closely related to viruses circulating in European pigs. J Gen Virol 2001;82:1397-1406. [PubMed]
82. Gundogdu Z, Seyhogullari M. Lymphadenitis as a Rare Side Effect of H1N1 Vaccine in a Child. Case Report Med 2010;2010:459543. [PubMed]
83. Gupta RK, George R, Nguyen-Van-Tam JS. Bacterial pneumonia and pandemic influenza planning. Emerg Infect Dis 2008;14:1187-1192. [PubMed]
84. Hackett S, Hill L, Patel J, Ratnaraja N, Ifeyinwa A, Farooqi M, Nusgen U, Debenham P, Gandhi D, Makwana N, Smit E, Welch S. Clinical characteristics of paediatric H1N1 admissions in Birmingham, UK. Lancet 2009;374:605. [PubMed]
85. Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009;361:1945-1952. [PubMed]
86. Hatchette TF, Bastien N, Berry J, Booth TF, Chernesky M, Couillard M, Drews S, Ebsworth A, Fearon M, Fonseca K, Fox J, Gagnon JN, Guercio S, Horsman G, Jorowski C, Kuschak T, Li Y, Majury A, Petric M, Ratnam S, Smieja M, Van Caeseele P. The limitations of point of care testing for pandemic influenza: what clinicians and public health professionals need to know. Can J Public Health 2009;100:204-207. [PubMed]
87. Hawkes M, Richardson SE, Ipp M, Schuh S, Adachi D, Tran D. Sensitivity of rapid influenza diagnostic testing for swine-origin 2009 a (H1N1) influenza virus in children. Pediatrics 2010;125:e639-644. [PubMed]
88. Hay AJ, Gregory V, Douglas AR, Lin YP. The evolution of human influenza viruses. Philos Trans R Soc Lond B Biol Sci 2001;356:1861-1870. [PubMed]
89. Hewagama S, Walker SP, Stuart RL, Gordon C, Johnson PD, Friedman ND, O'Reilly M, Cheng AC, Giles ML. 2009 H1N1 influenza A and pregnancy outcomes in Victoria, Australia. Clin Infect Dis 2010;50:686-690. [PubMed]
90. Heymann A, Chodick G, Reichman B, Kokia E, Laufer J. Influence of school closure on the incidence of viral respiratory diseases among children and on health care utilization. Pediatr Infect Dis J 2004;23:675-677. [PubMed]
91. Hollingsworth TD, Ferguson NM, Anderson RM. Frequent travelers and rate of spread of epidemics. Emerg Infect Dis 2007;13:1288-1294. [PubMed]
92. Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 2005;3:e300. [PubMed]
93. Hsieh YC, Wu TZ, Liu DP, Shao PL, Chang LY, Lu CY, Lee CY, Huang FY, Huang LM. Influenza pandemics: past, present and future. J Formos Med Assoc 2006;105:1-6. [PubMed]
94. Hu JJ, Kao CL, Lee PI, Chen CM, Lee CY, Lu CY, Huang LM. Clinical features of influenza A and B in children and association with myositis. J Microbiol Immunol Infect 2004;37:95-98. [PubMed]
95. Huang SM, Chen CC, Chiu PC, Cheng MF, Lai PH, Hsieh KS. Acute necrotizing encephalopathy of childhood associated with influenza type B virus infection in a 3-year-old girl. J Child Neurol 2004;19:64-67. [PubMed]
96. Hurt AC, Baas C, Deng YM, Roberts S, Kelso A, Barr IG. Performance of influenza rapid point-of-care tests in the detection of swine lineage A(H1N1) influenza viruses. Influenza Other Respi Viruses 2009;3:171-176. [PubMed]
97. Inglesby TV, Nuzzo JB, O'Toole T, Henderson DA. Disease mitigation measures in the control of pandemic influenza. Biosecur Bioterror 2006;4:366-375. [PubMed]
98. Jackson C, Mangtani P, Vynnycky E, Fielding K, Kitching A, Mohamed H, Roche A, Maguire H. School closures and student contact patterns. Emerg Infect Dis 2011;17:245-247. [PubMed]
99. Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, Sugerman DE, Druckenmiller JK, Ritger KA, Chugh R, Jasuja S, Deutscher M, Chen S, Walker JD, Duchin JS, Lett S, Soliva S, Wells EV, Swerdlow D, Uyeki TM, Fiore AE, Olsen SJ, Fry AM, Bridges CB, Finelli L. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med 2009;361:1935-1944. [PubMed]
100. Jamieson DJ, Honein MA, Rasmussen SA, Williams JL, Swerdlow DL, Biggerstaff MS, Lindstrom S, Louie JK, Christ CM, Bohm SR, Fonseca VP, Ritger KA, Kuhles DJ, Eggers P, Bruce H, Davidson HA, Lutterloh E, Harris ML, Burke C, Cocoros N, Finelli L, MacFarlane KF, Shu B, Olsen SJ. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet 2009;374:451-458. [PubMed]
101. Jeffries DJ. Viral hazards to and from health care workers. J Hosp Infect 1995;30 Suppl:140-155. [PubMed]
102. Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci U S A 2008;105:10967-10972. [PubMed]
103. Kandun IN, Tresnaningsih E, Purba WH, Lee V, Samaan G, Harun S, Soni E, Septiawati C, Setiawati T, Sariwati E, Wandra T. Factors associated with case fatality of human H5N1 virus infections in Indonesia: a case series. Lancet 2008;372:744-749. [PubMed]
104. Kandun IN, Wibisono H, Sedyaningsih ER, Yusharmen, Hadisoedarsuno W, Purba W, Santoso H, Septiawati C, Tresnaningsih E, Heriyanto B, Yuwono D, Harun S, Soeroso S, Giriputra S, Blair PJ, Jeremijenko A, Kosasih H, Putnam SD, Samaan G, Silitonga M, Chan KH, Poon LL, Lim W, Klimov A, Lindstrom S, Guan Y, Donis R, Katz J, Cox N, Peiris M, Uyeki TM. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med 2006;355:2186-2194. [PubMed]
105. Karre T, Maguire HF, Butcher D, Graepler A, Weed D, Wilson ML. Comparison of Becton Dickinson Directigen EZ Flu A+B test against the CDC real-time PCR assay for detection of 2009 pandemic influenza A/H1N1 virus. J Clin Microbiol 2010;48:343-344.[PubMed]
106. Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol 1989;63:4603-4608. [PubMed]
107. Kelso JK, Milne GJ, Kelly H. Simulation suggests that rapid activation of social distancing can arrest epidemic development due to a novel strain of influenza. BMC Public Health 2009;9:117. [PubMed]
108. Kitching A, Roche A, Balasegaram S, Heathcock R, Maguire H. Oseltamivir adherence and side effects among children in three London schools affected by influenza A(H1N1)v, May 2009 - an internet-based cross-sectional survey. Euro Surveill 2009;14:19287.[PubMed]
109. Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007;445:319-323. [PubMed]
110. Kok J, Blyth CC, Foo H, Patterson J, Taylor J, McPhie K, Ratnamohan VM, Iredell JR, Dwyer DE. Comparison of a rapid antigen test with nucleic acid testing during cocirculation of pandemic influenza A/H1N1 2009 and seasonal influenza A/H3N2. J Clin Microbiol 2010;48:290-291. [PubMed]
111. Komadina N, Roque V, Thawatsupha P, Rimando-Magalong J, Waicharoen S, Bomasang E, Sawanpanyalert P, Rivera M, Iannello P, Hurt AC, Barr IG. Genetic analysis of two influenza A (H1) swine viruses isolated from humans in Thailand and the Philippines. Virus Genes 2007;35:161-165. [PubMed]
112. Kumar A, Zarychanski R, Pinto R, Cook DJ, Marshall J, Lacroix J, Stelfox T, Bagshaw S, Choong K, Lamontagne F, Turgeon AF, Lapinsky S, Ahern SP, Smith O, Siddiqui F, Jouvet P, Khwaja K, McIntyre L, Menon K, Hutchison J, Hornstein D, Joffe A, Lauzier F, Singh J, Karachi T, Wiebe K, Olafson K, Ramsey C, Sharma S, Dodek P, Meade M, Hall R, Fowler RA. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA 2009;302:1872-1879. [PubMed]
113. Landry ML, Cohen S, Ferguson D. Comparison of Binax NOW and Directigen for rapid detection of influenza A and B. J Clin Virol 2004;31:113-115. [PubMed]
114. Landry ML, Cohen S, Ferguson D. Real-time PCR compared to Binax NOW and cytospin-immunofluorescence for detection of influenza in hospitalized patients. J Clin Virol 2008;43:148-151. [PubMed]
115. Larcombe PJ, Moloney SE, Schmidt PA. Pandemic (H1N1) 2009: a clinical spectrum in the general paediatric population. Arch Dis Child 2009. [PubMed]
116. Lemieux C, Brankston G, Gitterman L, Hirji Z, Gardam M. Questioning aerosol transmission of influenza. Emerg Infect Dis 2007;13:173-174; author reply 174-175. [PubMed]
117. Levi J, Inglesby T. Working Group on Pandemic Influenza Preparedness: joint statement in response to Department of Health and Human Services Pandemic Influenza Plan. Clin Infect Dis 2006;42:92-94. [PubMed]
118. Liem NT, Tung CV, Hien ND, Hien TT, Chau NQ, Long HT, Hien NT, Mai le Q, Taylor WR, Wertheim H, Farrar J, Khang DD, Horby P. Clinical features of human influenza A (H5N1) infection in Vietnam: 2004-2006. Clin Infect Dis 2009;48:1639-1646. [PubMed]
119. Ligon BL. Avian influenza virus H5N1: a review of its history and information regarding its potential to cause the next pandemic. Semin Pediatr Infect Dis 2005;16:326-335. [PubMed]
120. Lindsley WG, Blachere FM, Thewlis RE, Vishnu A, Davis KA, Cao G, Palmer JE, Clark KE, Fisher MA, Khakoo R, Beezhold DH. Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS One 2010;5:e15100. [PubMed]
121. Lister P, Reynolds F, Parslow R, Chan A, Cooper M, Plunkett A, Riphagen S, Peters M. Swine-origin influenza virus H1N1, seasonal influenza virus, and critical illness in children. Lancet 2009;374:605-607. [PubMed]
122. Loeb M, Dafoe N, Mahony J, John M, Sarabia A, Glavin V, Webby R, Smieja M, Earn DJ, Chong S, Webb A, Walter SD. Surgical Mask vs N95 Respirator for Preventing Influenza Among Health Care Workers: A Randomized Trial. JAMA 2009.[PubMed]
123. Louie JK, Guevara H, Boston E, Dahlke M, Nevarez M, Kong T, Schechter R, Glaser CA, Schnurr DP. Rapid influenza antigen test for diagnosis of pandemic (H1N1) 2009. Emerg Infect Dis 2010;16:824-826. [PubMed]
124. Louria DB, Blumenfeld HL, Ellis JT, Kilbourne ED, Rogers DE. Studies on influenza in the pandemic of 1957-1958. II. Pulmonary complications of influenza. J Clin Invest 1959;38:213-265. [PubMed]
125. Lu CY, Shao PL, Chang LY, Huang YC, Chiu CH, Hsieh YC, Lin TY, Huang LM. Immunogenicity and safety of a monovalent vaccine for the 2009 pandemic influenza virus A (H1N1) in children and adolescents. Vaccine 2010;28:5864-5870. [PubMed]
126. Luk J, Gross P, Thompson WW. Observations on mortality during the 1918 influenza pandemic. Clin Infect Dis 2001;33:1375-1378. [PubMed]
127. Madeley CR. Are point-of-care (POC) virological tests what is needed? Clin Microbiol Infect 2007;13:655-656. [PubMed]
128. Marshall CS, Styles K, Abraham G, Athan E. Management of potential laboratory exposure to avian influenza (H5N1) virus: implications for pandemic planning. Infect Control Hosp Epidemiol 2009;30:504-506. [PubMed]
129. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 2004;101:4620-4624. [PubMed]
130. Mermel LA. Preventing the spread of influenza A H1N1 2009 to health-care workers. Lancet Infect Dis 2009;9:723-724.[PubMed]
131. Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet 2010;375:1100-1108. [PubMed]
132. Montastruc JL, Durrieu G, Rascol O. Pandemrix degrees , (H1N1)v influenza and reported cases of narcolepsy. Vaccine 2011.[PubMed]
133. Morse SS, Garwin RL, Olsiewski PJ. Public health. Next flu pandemic: what to do until the vaccine arrives? Science 2006;314:929. [PubMed]
134. Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 2007;44:1084-1088. [PubMed]
135. Needham H. H5N1 in wild and domestic birds in Europe--remaining vigilant in response to an ongoing public health threat. Euro Surveill 2007;12:E071206 071201. [PubMed]
136. Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 2008;4:e1000012. [PubMed]
137. Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009;459:931-939. [PubMed]
138. Nguyen-Van-Tam JS, Openshaw PJ, Hashim A, Gadd EM, Lim WS, Semple MG, Read RC, Taylor BL, Brett SJ, McMenamin J, Enstone JE, Armstrong C, Nicholson KG. Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave (May-September 2009). Thorax 2010;65:645-651. [PubMed]
139. Nolan T, McVernon J, Skeljo M, Richmond P, Wadia U, Lambert S, Nissen M, Marshall H, Booy R, Heron L, Hartel G, Lai M, Basser R, Gittleson C, Greenberg M. Immunogenicity of a monovalent 2009 influenza A(H1N1) vaccine in infants and children: a randomized trial. JAMA 2010;303:37-46. [PubMed]
140. Noriega LM, Verdugo RJ, Araos R, Munita JM, Diaz V, Marcotti A, Perez J, Gonzalez P, Thompson L, Canals M, Hoppe A, Mounts AW, Vial PA. Pandemic influenza A (H1N1) 2009 with neurological manifestations, a case series. Influenza Other Respi Viruses 2010;4:117-120. [PubMed]
141. Oh CE, Lee J, Kang JH, Hong YJ, Kim YK, Cheong HJ, Ahn YJ, Kim SH, Lee HJ. Safety and immunogenicity of an inactivated split-virus influenza A/H1N1 vaccine in healthy children from 6 months to <18 years of age: a prospective, open-label, multi-center trial. Vaccine 2010;28:5857-5863. [PubMed]
142. Oseasohn R, Adelson L, Kaji M. Clinicopathologic study of thirty-three fatal cases of Asian influenza. N Engl J Med 1959;260:509-518. [PubMed]
143. Peebles PJ, Dhara R, Brammer L, Fry AM, Finelli L. Influenza-associated mortality among children - United States: 2007-2008. Influenza Other Respi Viruses 2010;5:25-31. [PubMed]
144. Peiris JS, Cheung CY, Leung CY, Nicholls JM. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol 2009;30:574-584. [PubMed]
145. Peiris JS, Poon LL, Guan Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol 2009;45:169-173. [PubMed]
146. Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 2004;363:617-619. [PubMed]
147. Perez-Padilla R, de la Rosa-Zamboni D, Ponce de Leon S, Hernandez M, Quinones-Falconi F, Bautista E, Ramirez-Venegas A, Rojas-Serrano J, Ormsby CE, Corrales A, Higuera A, Mondragon E, Cordova-Villalobos JA. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med 2009;361:680-689. [PubMed]
148. Pittet D. Improving adherence to hand hygiene practice: a multidisciplinary approach. Emerg Infect Dis 2001;7:234-240.[PubMed]
149. Pregliasco F, Puzelli S, Mensi C, Anselmi G, Marinello R, Tanzi ML, Affinito C, Zambon MC, Donatelli I. Influenza virological surveillance in children: the use of the QuickVue rapid diagnostic test. J Med Virol 2004;73:269-273. [PubMed]
150. Presanis AM, De Angelis D, Hagy A, Reed C, Riley S, Cooper BS, Finelli L, Biedrzycki P, Lipsitch M. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med 2009;6:e1000207. [PubMed]
151. Quach C, Newby D, Daoust G, Rubin E, McDonald J. QuickVue influenza test for rapid detection of influenza A and B viruses in a pediatric population. Clin Diagn Lab Immunol 2002;9:925-926. [PubMed]
152. Qureshi NR, Hien TT, Farrar J, Gleeson FV. The radiologic manifestations of H5N1 avian influenza. J Thorac Imaging 2006;21:259-264. [PubMed]
153. Rahman M, Kieke BA, Vandermause MF, Mitchell PD, Greenlee RT, Belongia EA. Performance of Directigen flu A+B enzyme immunoassay and direct fluorescent assay for detection of influenza infection during the 2004-2005 season. Diagn Microbiol Infect Dis 2007;58:413-418. [PubMed]
154. Rahman M, Vandermause MF, Kieke BA, Belongia EA. Performance of Binax NOW Flu A and B and direct fluorescent assay in comparison with a composite of viral culture or reverse transcription polymerase chain reaction for detection of influenza infection during the 2006 to 2007 season. Diagn Microbiol Infect Dis 2008;62:162-166. [PubMed]
155. Rambaut A, Holmes E. The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Curr 2009;1:RRN1003. [PubMed]
156. Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 "Spanish" influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A 1999;96:1651-1656. [PubMed]
157. Reid AH, Fanning TG, Janczewski TA, Lourens RM, Taubenberger JK. Novel origin of the 1918 pandemic influenza virus nucleoprotein gene. J Virol 2004;78:12462-12470. [PubMed]
158. Reid AH, Fanning TG, Janczewski TA, McCall S, Taubenberger JK. Characterization of the 1918 "Spanish" influenza virus matrix gene segment. J Virol 2002;76:10717-10723. [PubMed]
159. Reid AH, Fanning TG, Janczewski TA, Taubenberger JK. Characterization of the 1918 "Spanish" influenza virus neuraminidase gene. Proc Natl Acad Sci U S A 2000;97:6785-6790. [PubMed]
160. Rello J, Pop-Vicas A. Clinical review: primary influenza viral pneumonia. Crit Care 2009;13:235. [PubMed]
161. Rello J, Rodriguez A, Ibanez P, Socias L, Cebrian J, Marques A, Guerrero J, Ruiz-Santana S, Marquez E, Del Nogal-Saez F, Alvarez-Lerma F, Martinez S, Ferrer M, Avellanas M, Granada R, Maravi-Poma E, Albert P, Sierra R, Vidaur L, Ortiz P, Prieto del Portillo I, Galvan B, Leon-Gil C. Intensive care adult patients with severe respiratory failure caused by Influenza A (H1N1)v in Spain. Crit Care 2009;13:R148. [PubMed]
162. Russell KL, Ryan MA, Hawksworth A, Freed NE, Irvine M, Daum LT. Effectiveness of the 2003-2004 influenza vaccine among U.S. military basic trainees: a year of suboptimal match between vaccine and circulating strain. Vaccine 2005;23:1981-1985.[PubMed]
163. Ryan-Poirier KA, Katz JM, Webster RG, Kawaoka Y. Application of Directigen FLU-A for the detection of influenza A virus in human and nonhuman specimens. J Clin Microbiol 1992;30:1072-1075. [PubMed]
164. Sakai-Tagawa Y, Ozawa M, Tamura D, Le MQ, Nidom CA, Sugaya N, Kawaoka Y. Sensitivity of influenza rapid diagnostic tests to H5N1 and 2009 pandemic H1N1 viruses. J Clin Microbiol 2010;48:2872-2877. [PubMed]
165. Sambol AR, Abdalhamid B, Lyden ER, Aden TA, Noel RK, Hinrichs SH. Use of rapid influenza diagnostic tests under field conditions as a screening tool during an outbreak of the 2009 novel influenza virus: practical considerations. J Clin Virol 2010;47:229-233. [PubMed]
166. Sandora TJ, Smole SC, Lee GM, Chung S, Williams L, McAdam AJ. Test characteristics of commercial influenza assays for detecting pandemic influenza A (H1N1) in children. Pediatr Infect Dis J 2010;29:261-262. [PubMed]
167. Schotsaert M, De Filette M, Fiers W, Saelens X. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Rev Vaccines 2009;8:499-508. [PubMed]
168. Seale H, Corbett S, Dwyer DE, MacIntyre CR. Feasibility exercise to evaluate the use of particulate respirators by emergency department staff during the 2007 influenza season. Infect Control Hosp Epidemiol 2009;30:710-712. [PubMed]
169. Selvarangan R, Abel D, Hamilton M. Comparison of BD Directigen EZ RSV and Binax NOW RSV tests for rapid detection of respiratory syncytial virus from nasopharyngeal aspirates in a pediatric population. Diagn Microbiol Infect Dis 2008;62:157-161.[PubMed]
170. Sencer DJ, Millar JD. Reflections on the 1976 swine flu vaccination program. Emerg Infect Dis 2006;12:29-33. [PubMed]
171. Seto WH, Tsang D, Yung RW, Ching TY, Ng TK, Ho M, Ho LM, Peiris JS. Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 2003;361:1519-1520.[PubMed]
172. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature 2006;440:435-436. [PubMed]
173. Sickbert-Bennett EE, Weber DJ, Gergen-Teague MF, Sobsey MD, Samsa GP, Rutala WA. Comparative efficacy of hand hygiene agents in the reduction of bacteria and viruses. Am J Infect Control 2005;33:67-77. [PubMed]
174. Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 2007;24:1811-1820. [PubMed]
175. Smit M, Beynon KA, Murdoch DR, Jennings LC. Comparison of the NOW Influenza A & B, NOW Flu A, NOW Flu B, and Directigen Flu A+B assays, and immunofluorescence with viral culture for the detection of influenza A and B viruses. Diagn Microbiol Infect Dis 2007;57:67-70. [PubMed]
176. Stanekova Z, Vareckova E. Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development. Virol J 2010;7:351. [PubMed]
177. Stelzer-Braid S, Oliver BG, Blazey AJ, Argent E, Newsome TP, Rawlinson WD, Tovey ER. Exhalation of respiratory viruses by breathing, coughing, and talking. J Med Virol 2009;81:1674-1679. [PubMed]
178. Sypsa V, Hatzakis A. School closure is currently the main strategy to mitigate influenza A(H1N1)v: a modeling study. Euro Surveill 2009;14. [PubMed]
179. Tanaka T, Nakajima K, Murashima A, Garcia-Bournissen F, Koren G, Ito S. Safety of neuraminidase inhibitors against novel influenza A (H1N1) in pregnant and breastfeeding women. CMAJ 2009;181:55-58. [PubMed]
180. Tang JW, Li Y. Transmission of influenza A in human beings. Lancet Infect Dis 2007;7:758; author reply 761-753. [PubMed]
181. Tang JW, Shetty N, Lam TT, Hon KL. Emerging, novel, and known influenza virus infections in humans. Infect Dis Clin North Am 2010;24:603-617. [PubMed]
182. Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 2006;12:15-22. [PubMed]
183. Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature 2005;437:889-893. [PubMed]
184. Taylor J, McPhie K, Druce J, Birch C, Dwyer DE. Evaluation of twenty rapid antigen tests for the detection of human influenza A H5N1, H3N2, H1N1, and B viruses. J Med Virol 2009;81:1918-1922. [PubMed]
185. Tellier R. Review of aerosol transmission of influenza A virus. Emerg Infect Dis 2006;12:1657-1662. [PubMed]
186. Tellier R. Transmission of influenza A in human beings. Lancet Infect Dis 2007;7:759-760; author reply 761-753. [PubMed]
187. Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface 2009;6 Suppl 6:S783-790.[PubMed]
188. To KF, Chan PK, Chan KF, Lee WK, Lam WY, Wong KF, Tang NL, Tsang DN, Sung RY, Buckley TA, Tam JS, Cheng AF. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J Med Virol 2001;63:242-246. [PubMed]
189. Toner E, Waldhorn R, Maldin B, Borio L, Nuzzo JB, Lam C, Franco C, Henderson DA, Inglesby TV, O'Toole T. Hospital preparedness for pandemic influenza. Biosecur Bioterror 2006;4:207-217. [PubMed]
190. Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen VC, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 2004;350:1179-1188. [PubMed]
191. Uyeki TM. Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin Infect Dis 2009;49:279-290. [PubMed]
192. Uyeki TM, Chong YH, Katz JM, Lim W, Ho YY, Wang SS, Tsang TH, Au WW, Chan SC, Rowe T, Hu-Primmer J, Bell JC, Thompson WW, Bridges CB, Cox NJ, Mak KH, Fukuda K. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg Infect Dis 2002;8:154-159. [PubMed]
193. Uyeki TM, Prasad R, Vukotich C, Stebbins S, Rinaldo CR, Ferng YH, Morse SS, Larson EL, Aiello AE, Davis B, Monto AS. Low sensitivity of rapid diagnostic test for influenza. Clin Infect Dis 2009;48:e89-92. [PubMed]
194. Vajo Z, Tamas F, Sinka L, Jankovics I. Safety and immunogenicity of a 2009 pandemic influenza A H1N1 vaccine when administered alone or simultaneously with the seasonal influenza vaccine for the 2009-10 influenza season: a multicentre, randomised controlled trial. Lancet 2010;375:49-55. [PubMed]
195. Van der Sluijs KF, van der Poll T, Lutter R, Juffermans NP, Schultz MJ. Bench-to-bedside review: bacterial pneumonia with influenza - pathogenesis and clinical implications. Crit Care 2010;14:219. [PubMed]
196. Van der Vliet D, Pepin S, Lambert M, Fauchoux N, Donazzolo Y, Dupuy M, Dakowski C, Denis M. Similar immunogenicity and safety of the A/H1N1 2009 pandemic influenza strain when used as a monovalent or a trivalent vaccine. Hum Vaccin 2010;6.[PubMed]
197. Van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science 2006;312:399. [PubMed]
198. Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin Infect Dis 2009;49:1090-1093. [PubMed]
199. Velasco JM, Montesa-Develos ML, Jarman RG, Lopez MN, Gibbons RV, Valderama MT, Yoon IK. Evaluation of QuickVue influenza A+B rapid test for detection of pandemic influenza A/H1N1 2009. J Clin Virol 2010;48:120-122. [PubMed]
200. Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS, Guan Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 2010;328:1529. [PubMed]
201. Wallensten A, Oliver I, Lewis D, Harrison S. Compliance and side effects of prophylactic oseltamivir treatment in a school in South West England. Euro Surveill 2009;14:19285. [PubMed]
202. Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol 1993;67:5585-5594. [PubMed]
203. Wang YH, Huang YC, Chang LY, Kao HT, Lin PY, Huang CG, Lin TY. Clinical characteristics of children with influenza A virus infection requiring hospitalization. J Microbiol Immunol Infect 2003;36:111-116. [PubMed]
204. Webb SA, Pettila V, Seppelt I, Bellomo R, Bailey M, Cooper DJ, Cretikos M, Davies AR, Finfer S, Harrigan PW, Hart GK, Howe B, Iredell JR, McArthur C, Mitchell I, Morrison S, Nichol AD, Paterson DL, Peake S, Richards B, Stephens D, Turner A, Yung M. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med 2009;361:1925-1934.[PubMed]
205. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992;56:152-179. [PubMed]
206. Weinberg A, Walker ML. Evaluation of three immunoassay kits for rapid detection of influenza virus A and B. Clin Diagn Lab Immunol 2005;12:367-370. [PubMed]
207. WHO. Recommended composition of influenza virus vaccines for use in the 2009 southern hemisphere influenza season. WHO Wkly Epidemiol Rec 2008;41:365-372. [PubMed]
208. WHO. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 2008;14:e1.[PubMed]
209. WHO. Interim WHO guidance for the surveillance of human infection with swine influenza A(H1N1) virus Available at:http://www.wpro.who.int/NR/rdonlyres/0F345099-6975-48DD-9B3F-F1C0DEB4255F/0/WHO_case_definition_swine_flu_2009_04_29.pdf. Accessed on February 25, 2011.
210. WHO. Recommended composition of influenza virus vaccines for use in the 2009–2010 influenza season (northern hemisphere winter). WHO Wkly Epidemiol Rec 2009;9:65-76. [PubMed]
211. WHO. Recommended composition of influenza virus vaccines for use in the 2010 influenza season (southern hemisphere winter). WHO Wkly Epidemiol Rec 2009;41:421-436. [PubMed]
212. WHO. WHO 2009 treatment guide. Available at:http://www.who.int/csr/resources/publications/swineflu/h1n1_guidelines_pharmaceutical_mngt.pdf. Accessed on August 28, 2009.
213. WHO. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. Available at:http://www.who.int/csr/disease/avian_influenza/country/en/. Accessed on February 25, 2011.
214. WHO. Recommended viruses for influenza vaccines for use in the 2010-2011 northern hemisphere influenza season. WHO Wkly Epidemiol Rec 2010;10:81-92. [PubMed]
215. WHO. Health topics: Avian influenza. Available at: http://www.who.int/topics/avian_influenza/en/. Accessed on 26 January, 2011.
216. Worobey M, Rambaut A, Pybus OG, Robertson DL. Questioning the evidence for genetic recombination in the 1918 "Spanish flu" virus. Science 2002;296:211 discussion 211. [PubMed]
217. Wunderink RG. Influenza and bacterial pneumonia--constant companions. Crit Care 2010;14:150. [PubMed]
218. Yang W, Elankumaran S, Marr LC. Concentrations and size distributions of airborne influenza A viruses measured indoors at a health centre, a day-care centre and on aeroplanes. J R Soc Interface 2011. [PubMed]
219. Yu H, Gao Z, Feng Z, Shu Y, Xiang N, Zhou L, Huai Y, Feng L, Peng Z, Li Z, Xu C, Li J, Hu C, Li Q, Xu X, Liu X, Liu Z, Xu L, Chen Y, Luo H, Wei L, Zhang X, Xin J, Guo J, Wang Q, Yuan Z, Zhang K, Zhang W, Yang J, Zhong X, Xia S, Li L, Cheng J, Ma E, He P, Lee SS, Wang Y, Uyeki TM, Yang W. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One 2008;3:e2985. [PubMed]
220. Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 1998;351:467-471.[PubMed]
Figure 1. 1A-1B. A negatively-stained transmission electron micrograph (TEM) of the Hong Kong H3N2 influenza virus responsible for the influenza pandemic of 1968-1969. This pandemic was estimated to have infected 50,000,000 people in the United States alone, resulting in about 33,000 deaths. A protein coat or capsid surrounds each individual virus with the ‘hairy’ hemagglutinin (HA) and neuraminidase (NA) spikes being clearly seen on the outer envelope surface in 1A (more easily seen in the schematic in 1B). These surface HA and NA proteins define the influenza A subtypes, such as H1N1, H2N2, H3N2 and H5N1. Closely matching these proteins within the seasonal influenza vaccines is important to induce specific protective host immunity against a particular influenza virus subtype. [Photo credit: Centers for Disease Control Public Health Image Library (CDC PHIL): http://phil.cdc.gov/phil/home.asp,
Figure 2: Schematic diagram showing the reassortment history of human pandemic influenza strains. Each influenza gene segment is represented by a colored, horizontal bar. The 1918 ‘Spanish flu’ influenza and classic swine influenza probably originated from an avian influenza virus population at some point in the past, but arrows for their origins have been omitted here because the exact species-crossing events cannot be defined for certain and remain controversial. The reassortment events generating the H3N2 Hong Kong flu pandemic strain have been simplified here due to space constraints (181).
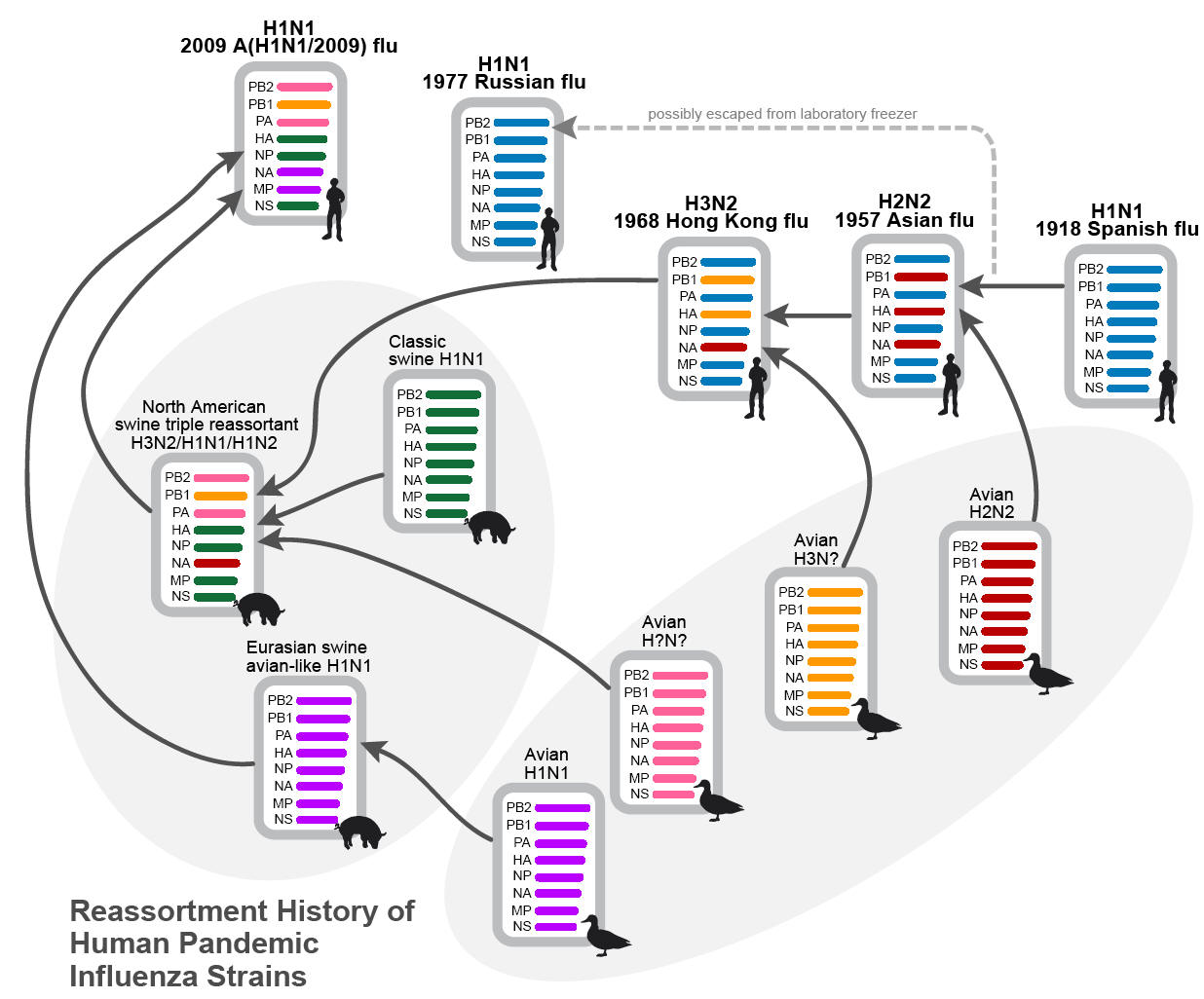
Figure 3. 3A.3B. Embyonated chicken egg culture for influenza viruses is no longer used in routine hospital diagnostic laboratories, though reference laboratories may still retain this capability. This older photograph from 1968 shows a laboratory worker inoculating 10-day-old embryonated chicken eggs with specimens suspected of containing influenza virus. 3B. Examining a viral culture flask of Madin-Darby Canine Kidney epithelial cells (MDCK) for any signs of influenza virus growth. The level of personal protective equipment (PPE) worn for such a procedure will depend on the lethality of that particular influenza virus subtype. In this case, full PPE would be worn when handling avian A(H5N1) virus, which has a 50-60% mortality for human infections. [Photo credit: CDC PHIL:http://phil.cdc.gov/phil/home.asp]
A  B
B 
Figure 4. An example of a real-time, reverse transcription polymerase chain reaction (RT-PCR) for pandemic A(H1N1) influenza virus in routine use from a hospital molecular diagnostic laboratory, showing a positive control trace (magenta, CP value 31.31) and four positive clinical samples (with CP values ranging from 25-36). The CP value is the number of PCR cycles required to cross a certain signal threshold indicating a positive result. These molecular tests are performed in specialized diagnostic laboratories with highly-skilled staff and offer greater sensitivity than the rapid test kits, though they take far longer (about 3-4 hours). [Photo credit: Courtesy of Professor Evelyn SC Koay, Molecular Diagnosis Centre, National University Hospital, Singapore.]
Figure 5. An example of an FDA-approved, rapid point-of-care test – the QuickVue Influenza A+B test (Quidel Corp Inc., San Diego, CA, USA). This is just an example of these kits, which sacrifice test sensitivity (which can be anything between 20%-90%, depending on the particular study cited) for a rapid test result (of about 10-15 minutes). There are many others on the market and the authors do not in anyway endorse this particular kit. [Photo credit: Julian W Tang]
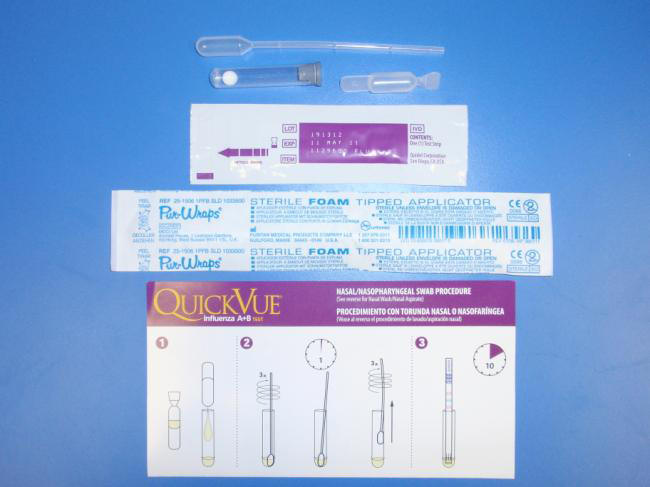
Figure 6. Influenza vaccines are typically given intramuscularly into the deltoid muscle in older children, as shown here. [Photo credit: Centers for Disease Control Public Health Image Library (CDC PHIL):http://phil.cdc.gov/phil/home.asp]
Table 1. Recommended Antiviral Dosing Regimens (For Both Oseltamivir And Zanamivir) For The Treatment Or Prophylaxis Of Pandemic Influenza A(H1N1).
| Recommended dose of oseltamivir for treatment (5 days) | Recommended dose of oseltamivir for prophylaxis (for at least 10 days)* | Recommended dose of zanamivir for treatment | Recommended dose of zanamivir for prophylaxis | |
|---|---|---|---|---|
Adults |
75 mg twice daily |
75 mg once daily |
Two 5-mg inhalations (10 mg total) twice per day |
Two 5-mg inhalation (10 mg total) once per day |
Children < 12 months |
|
|
Two 5-mg inhalations (10 mg total) twice per day (age, 7 years or older) |
Two 5-mg inhalation (10 mg total) once per day (age, 5 years or older) |
< 3 months |
12 mg twice daily |
Not recommended unless critically ill due to limited data in this age group |
|
|
3-5 months |
20 mg twice daily |
20 mg once daily |
|
|
6-11 months |
25 mg twice daily |
25 mg once daily |
|
|
Children > 12 months |
|
|
|
|
15 kg or less |
60 mg/day, divided into 2 doses |
30 mg once daily |
|
|
16-23 kg |
90 mg/day divided into 2 doses |
45 mg once daily |
|
|
24-40 kg |
120 mg/day, divided into 2 doses |
60 mg once daily |
|
|
> 40 kg |
150 mg/day, divided into 2 doses |
75 mg once daily |
|
|
*as recommended in the manufacturer's fact sheet on oseltamivir for healthcare providers at: http://www.cdc.gove/h1n1flu/EU/tamiflu-hcp.pdf
Readers Are Encouraged To Check For The Latest Updates From The CDC Website: http://www.cdc.gov/h1n1flu/recommendations.htm, as Some of These Recommendations May Have Changed by the Time
This Chapter Goes to Print. [Table adapted from reference (181)]
Table 2: Influenza Vaccines Recommended by the Advisory Committee on Immunization Practices (ACIP) for Different Age Groups — United States, 2010–11 Season (from: the CDC’s Seasonal Influenza Website at: http://www.cdc.gov/flu/protect/vaccine/vaccines.htm)
| Vaccine | Trade Name | Manufacturer | Presentation | Mercury content (mcg Hg/0.5 mL dose) | Age group | No. of doses | Route |
|---|---|---|---|---|---|---|---|
TIV* |
Fluzone |
Sanofi Pasteur |
0.25 mL prefilled syringe |
0 |
6-35 mos |
1 or 2† |
Intramuscular§ |
|
|
|
0.5 mL prefilled syringe |
0 |
36 mos and older |
1 or 2† |
Intramuscular§ |
|
|
|
0.5 mL vial |
0 |
36 mos and older |
1 or 2† |
Intramuscular§ |
|
|
|
5.0 mL multidose vial |
25 |
6 mos and older |
1 or 2† |
Intramuscular§ |
TIV |
Fluvirin |
Novartis Vaccines and Diagnostics |
5.0 mL multidose vial |
25 |
4 yrs and older |
1 or 2† |
Intramuscular§ |
|
|
|
0.5 mL prefilled syringe |
Less than 1.0 |
|
|
|
TIV |
Agriflu |
Novartis Vaccines and Diagnostics |
0.5 mL prefilled syringe |
0 |
18 yrs and older |
1 |
Intramuscular§ |
TIV |
Fluarix |
GlaxoSmithKline Biologicals |
0.5 mL prefilled syringe |
0 |
3 yrs and older |
1 or 2† |
Intramuscular§ |
TIV |
FluLaval |
ID Biomedical Corp. of Quebec, a subsidiary of GlaxoSmithKline |
5.0 mL multidose vial |
25 |
18yrs and older |
1 |
Intramuscular§ |
TIV |
Afluria¶ |
CSL Biotherapies |
0.5 mL prefilled syringe |
0 |
9 yrs and older |
1 |
Intramuscular§ |
TIV High Dose** |
Fluzone High-Dose |
Sanofi Pasteur |
0.5 mL prefilled syringe |
0 |
65 yrs and older |
1 |
Intramuscular§ |
LAIV†† |
FluMist§§ |
MedImmune |
0.2 mL sprayer, divided dose |
0 |
2-49 yrs |
1 or 2† |
Intranasal |
* Trivalent inactivated vaccine.
§Children aged 6 months--8 years who did not receive at least 1 dose of an influenza A (H1N1) 2009 monovalent vaccine, who have never received a seasonal influenza vaccine before, or who were vaccinated for the first time with the seasonal 2009--10 seasonal vaccine but who received only 1 dose should receive 2 doses of the 2010--11 influenza vaccine formula, spaced 4 or more weeks apart.
§For adults and older children, the recommended site of vaccination is the deltoid muscle. The preferred site for infants and young children is the anterolateral aspect of the thigh.
¶Afluria (CSL Biotherapies) is approved in the United States by the Food and Drug Administration for use in persons aged 6 months and older. However, the Advisory Committee on Immunization Practices recommends that the 2010--11 formulation of Afluria not be administered to children aged 6 months--8 years because of an increased frequency of fever or febrile seizures reported among young children (mostly children aged younger than 5 years) who received a similar vaccine in Australia in 2010. Therefore, another age-appropriate, licensed seasonal influenza vaccine formulation should be used for prevention of influenza in children aged 6 months--8 years. If no other age-appropriate, licensed seasonal influenza vaccine is available for a child aged 5--8 years who has a medical condition that increases the child's risk for influenza complications, Afluria may be given. Providers should discuss with the parents or caregivers the benefits and risks of influenza vaccination with Afluria before administering this vaccine. See second footnote above for dose information when administering Afluria to children aged 5--8 years.
** Trivalent inactivated vaccine high dose. A 0.5-mL dose contains 60 mcg each of A/California/7/2009 (H1N1)-like, A/Perth/16/2009 (H3N2)-like, and B/Brisbane/60/2008-like antigens.
††Live attenuated influenza vaccine.
§§ FluMist is shipped refrigerated and stored in the refrigerator at 36°F--46°F (2°C--8°C) after arrival in the vaccination clinic. The dose is 0.2 mL divided equally between each nostril. Health-care providers should consult the medical record, when available, to identify children aged 2--4 years with asthma or recurrent wheezing that might indicate asthma. In addition, to identify children who might be at greater risk for asthma and possibly at increased risk for wheezing after receiving LAIV, parents or caregivers of children aged 2--4 years should be asked: "In the past 12 months, has a health-care provider ever told you that your child had wheezing or asthma?" Children whose parents or caregivers answer "yes" to this question and children who have asthma or who had a wheezing episode noted in the medical record within the past 12 months should not receive FluMist.
Table 3: Non-Pharmaceutical Public Health Interventions (2)
| Human surveillance | Community Restrictions | Patient Management | Contact Management | ||||
|---|---|---|---|---|---|---|---|
|
|
|
|
1Gowns, gloves and protective eye covers
2Separating exposed individuals from others
3Voluntary sequestration of healthy persons to avoid exposure
4Exit and entry screening, travel advisories
Box 1. Recommendations for the receipt of the seasonal influenza vaccine, 2010 (from CDC website: Persons for Whom Annual Vaccination is Recommended:http://www.cdc.gov/flu/professionals/acip/persons.htm
| All persons aged 6 months and older should be vaccinated annually.
Protection of persons at higher risk for influenza-related complications should continue to be a focus of vaccination efforts as providers and programs transition to routine vaccination of all persons aged 6 months and older. When vaccine supply is limited, vaccination efforts should focus on delivering vaccination to persons who: o are aged 6 months--4 years (59 months); o are aged 50 years and older; o have chronic pulmonary (including asthma), cardiovascular (except hypertension), renal, hepatic, neurologic, hematologic, or metabolic disorders (including diabetes mellitus); o are immunosuppressed (including immunosuppression caused by medications or by human immunodeficiency virus); o are or will be pregnant during the influenza season; o are aged 6 months--18 years and receiving long-term aspirin therapy and who therefore might be at risk for experiencing Reye syndrome after influenza virus infection; o are residents of nursing homes and other chronic-care facilities; o are American Indians/Alaska Natives; o are morbidly obese (body-mass index is 40 or greater); o are health-care personnel; o are household contacts and caregivers of children aged younger than 5 years and adults aged 50 years and older, with particular emphasis on vaccinating contacts of children aged younger than 6 months; and o are household contacts and caregivers of persons with medical conditions that put them at higher risk for severe complications from influenza. |
Box 2: Seasonal influenza vaccine composition before and after the 2009 A(H1N1) influenza pandemic.

Box 3. Example of a combined clinical and laboratory case definition for pandemic influenza A(H1N1) from the World Health Organization website (209).

Clinical Case Section
Courtesy of Dr. Nandini Shetty, University College London Hospitals, London, UK
Figure A: Serial chest X-ray changes for Patient A in a fatal pandemic influenza A(H1N1) infection. Note the rapid, progression of bilateral infiltration with widespread consolidation, in keeping with an aggressive viral pneumonia with a likely (though treated) bacterial secondary pneumonia
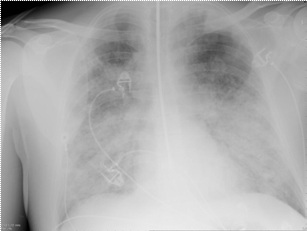 A On admission (Day 1)
A On admission (Day 1)
 B Day 3 (on ICU)
B Day 3 (on ICU)
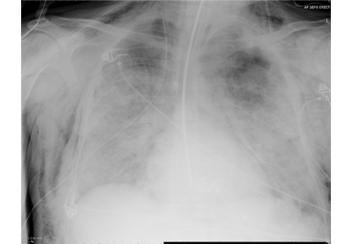 C Day 4 (on ICU)
C Day 4 (on ICU)
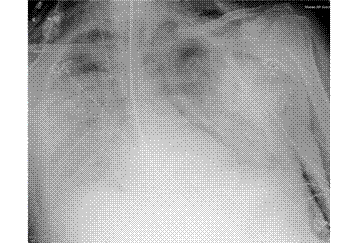 D Day 5 (one day before death)
D Day 5 (one day before death)
Figure B: Serial chest X-ray changes for Patient B who was 29 weeks pregnant at the time of her pandemic influenza A(H1N1) infection. Note the gradual resolution of the pulmonary infiltrates and consolidation. The emergency delivery of her baby allowed her respiratory function to improve significantly.
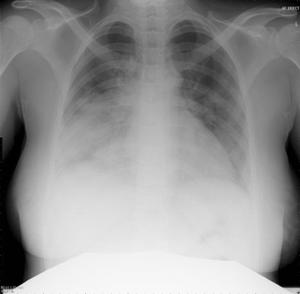 A On Day 1 of admission
A On Day 1 of admission
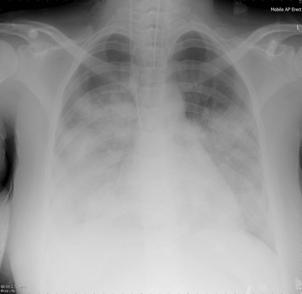 B On Day 2 of admission
B On Day 2 of admission
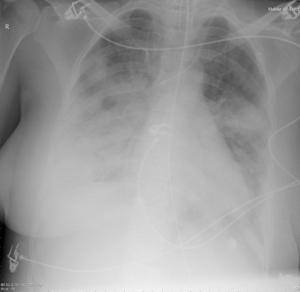 C On Day 9 of admission
C On Day 9 of admission
Figure C: Serial chest X-ray changes for Patient C with a persistent A(H1N1) infection. In this recovering case, note the gradually resolution of the pulmonary infiltrates and consolidation.
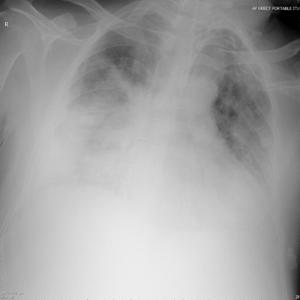 A On Day 6 of admission
A On Day 6 of admission
 B On Day 11
B On Day 11
What's New
Madhi SA, et al. Influenza Vaccination of Pregnant Women and Protection of Their Infants. New Engl J Med 2014;371:918-931.
Diaz Granados CA, et al. Efficacy of high-dose versus standard-dose influenza vaccine in older adults. N Engl J Med 2014:371:635.
South East Asia Infectious Disease Clinical Research Network. Effect of double dose oseltamivir on clinical and virological outcomes in children and adults admitted to hospital with severe influenza: double blind randomised controlled trial. BMJ 2013;346:f3039 (pages 1-16).
van Asten L, et al. Mortality Attributable to 9 Common Infections: Significant Effect of Influenza A, Respiratory Syncytial Virus, Influenza B, Norovirus, and Parainfluenza in Elderly Persons. J Infect Dis 2012;206:628.
Graitcer SB, et al. Characteristics of Patients with Oseltamivir-Resistant Pandemic (H1N1) 2009, United States. Emerg Infect Dis 2011;17:255-257.
Rizzo C, et al. Pandemic Influenza and Pneumonia Due to Legionella pneumophila: A Frequently Underestimated Coinfection.Clin Infect Dis. 2010 Jul 1;51:115.
Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza. Clinical Aspects of Pandemic 2009 Influenza A (H1N1) Virus Infection. New Engl J Med 2010;362:1708-1719.
Ang B, et al. Surgical masks for protection of health care personnel against pandemic novel swine-origin influenza A (H1N1)-2009: results from an observational study. Clin Infect Dis 2010; 50 :1011-4.
Li CC, et al. Correlation of Pandemic (H1N1) 2009 Viral Load with Disease Severity and Prolonged Viral Shedding in Children. Emerg Infect Dis 2010;16:1265-1272.
Creanga AA, Johnson TF, et al. Severity of 2009 Pandemic Influenza A (H1N1) Virus Infection in Pregnant Women. Obstet Gynecol. 2010 Apr;115:717-26.
De Serres G, et al. Contagious Period for Pandemic (H1N1) 2009. Emerg Infect Dis. 2010 May;16:783-8.
Nguyen JT, Hoopes JD, et al. Triple Combination of Oseltamivir, Amantadine, and Ribavirin Displays Synergistic Activity Against Multiple Influenza Virus Strains in vitro. Antimicrob Agents Chemother. 2009 Oct;53(10):4115-26.
Cauchemez S, et al. Household Transmission of 2009 Pandemic Influenza A (H1N1) Virus in the United States. N Engl J Med 2009;361 (27):2619-2627.
CDC recommends against the use of amantadine and rimantadine 2006
Webster RG. H5N1 Outbreaks and Enzootic Influenza. Emerg Infect Dis, Jan 2006.
GUIDED MEDLINE SEARCH FOR:
Review articles
World Health Organization. Avian influenza: assessing the pandemic threat
Webster RG, Guan Y, Peiris M, Chen H. H5N1 Influenza Continues To Circulate and Change. Microbe 2006;12:559-565.
Woolhouse MEJ. Where Do Emerging Pathogens Come from? Microbe 2006;1(11):511-515.
Tosh PK, Boyce TG, Poland GA, Vaccine Research Group. Flu myths: dispelling the myths associated with live attenuated influenza vaccine. Mayo Clin Proc. 2008; 83(1):77-84.
Management of Influenza - An Update for 2009.
ACIP: Prevention and Control of Influenza with Vaccines. MMWR, 2010.
GUIDED MEDLINE SEARCH FOR RECENT REVIEWS
History
Rosenberg CE. Siting Epidemic Disease: 3 Centuries of American History. J Infect Dis 2008;197 (Suppl 1):S4-S6.
Schultz MG, Morens DM. Charles-Jules-Henri Nicolle: Contributions to Typhus and Influenza. Emerging Infectious Diseases 2009;15(9):1520-1522.
Trilla A, et al. The 1918 "Spanish flu" in Spain. Clin Infect Dis. 2008 Sep 1;47:668-73.
Zimmer SM, et al. Historical Perspective -- Emergence of Influenza A (H1N1) Viruses. N Engl J Med 2009;361:279-285.
Morens DM, et al. The Persistent Legacy of the 1918 Influenza Virus. N Engl J Med 2009;361 (3):225-229.
Kilbourne ED. Influenza Pandemics of 20th Century. Emerg Infect Dis, Jan 2006.
Taubenberger JK, Morens DM. 1918 Influenza: the Mother of All Pandemics. Emerg Infect Dis, Jan 2006.
Krause R. The Swine Flu Episode and the Fog of Epidemics. Emerg Infect Dis, Jan 2006.
Billings M, Schneider J. 2007. The Influenza Pandemic of 1918.
Morens DM, Fauci AS. The 1918 Influenza Pandemic: Insights for the 21st Century. Journal of Infectious Diseases, April 2007.
GUIDED MEDLINE SEARCH FOR HISTORICAL ASPECTS
Table of Contents
- Virology
- Epidemiology
- Clinical Manifestations
- Laboratory Diagnosis
- Antimicrobial Therapy
- Vaccines
- Prevention
- Conclusions
- Clinical Case Section
